Identification of stably expressed reference genes for expression studies in Arabidopsis thaliana using mass spectrometry-based label-free quantification
- PMID: 36247637
- PMCID: PMC9557097
- DOI: 10.3389/fpls.2022.1001920
Identification of stably expressed reference genes for expression studies in Arabidopsis thaliana using mass spectrometry-based label-free quantification
Abstract
Arabidopsis thaliana has been used regularly as a model plant in gene expression studies on transcriptional reprogramming upon pathogen infection, such as that by Pseudomonas syringae pv. tomato DC3000 (Pst DC3000), or when subjected to stress hormone treatments including jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) has been extensively employed to quantitate these gene expression changes. However, the accuracy of the quantitation is largely dependent on the stability of the expressions of reference genes used for normalization. Recently, RNA sequencing (RNA-seq) has been widely used to mine stably expressed genes for use as references in RT-qPCR. However, the amplification step in RNA-seq creates an intrinsic bias against those genes with relatively low expression levels, and therefore does not provide an accurate quantification of all expressed genes. In this study, we employed mass spectrometry-based label-free quantification (LFQ) in proteomic analyses to identify those proteins with abundances unaffected by Pst DC3000 infection. We verified, using RT-qPCR, that the levels of their corresponding mRNAs were also unaffected by Pst DC3000 infection. Compared to commonly used reference genes for expression studies in A. thaliana upon Pst DC3000 infection, the candidate reference genes reported in this study generally have a higher expression stability. In addition, using RT-qPCR, we verified that the mRNAs of the candidate reference genes were stably expressed upon stress hormone treatments including JA, SA, and ABA. Results indicated that the candidate genes identified here had stable expressions upon these stresses and are suitable to be used as reference genes for RT-qPCR. Among the 18 candidate reference genes reported in this study, many of them had greater expression stability than the commonly used reference genes, such as ACT7, in previous studies. Here, besides proposing more appropriate reference genes for Arabidopsis expression studies, we also demonstrated the capacity of mass spectrometry-based LFQ to quantify protein abundance and the possibility to extend protein expression studies to the transcript level.
Keywords: RT-qPCR; abscisic acid; expression study; jasmonic acid; label-free quantification; pathogen infection; reference gene; salicylic acid.
Copyright © 2022 Cheng, Ku, Cheung and Lam.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
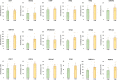



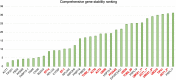

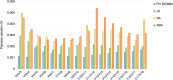
References
-
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496, PMID: - DOI - PubMed
-
- Arefian M., Vessal S., Malekzadeh-Shafaroudi S., Siddique K. H. M., Bagheri A. (2019). Comparative proteomics and gene expression analyses revealed responsive proteins and mechanisms for salt tolerance in chickpea genotypes. BMC Plant Biol. 19:300. doi: 10.1186/s12870-019-1793-z, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

