Identification of cystic fibrosis transmembrane conductance regulator as a prognostic marker for juvenile myelomonocytic leukemia via the whole-genome bisulfite sequencing of monozygotic twins and data mining
- PMID: 36247890
- PMCID: PMC9561505
- DOI: 10.21037/tp-22-381
Identification of cystic fibrosis transmembrane conductance regulator as a prognostic marker for juvenile myelomonocytic leukemia via the whole-genome bisulfite sequencing of monozygotic twins and data mining
Abstract
Background: Linked deoxyribonucleic acid (DNA) hypermethylation investigations of promoter methylation levels of candidate genes may help to increase the progressiveness and mortality rates of juvenile myelomonocytic leukemia (JMML), which is a unique myelodysplastic/myeloproliferative neoplasm caused by excessive monocyte and granulocyte proliferation in infancy/early childhood. However, the roles of hypermethylation in this malignant disease are uncertain.
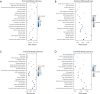
Methods: Bone marrow samples from a JMML patient and peripheral blood samples from a healthy monozygotic twin and an unrelated healthy donor were collected with the informed consent of the participant's parents. Whole-genome bisulfite sequencing (WGBS) was then performed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to analyze specific differentially methylated region (DMG) related genes. The target genes were screened with Cytoscape and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), which are gene/protein interaction databases. A data mining platform was used to examine the expression level data of the healthy control and JMML patient tissues in Gene Expression Omnibus data sets, and a survival analysis was performed for all the JMML patients.
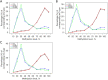
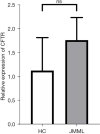
Results: The STRING analysis revealed that the red node [i.e., the cystic fibrosis transmembrane conductance regulator (CFTR)] was the gene of interest. The gene-expression microarray data set analysis suggested that the CFTR expression levels did not differ significantly between the JMML patients and healthy controls (P=0.81). A statistically significant difference was observed in the CFTR promoter methylation level but not in the CFTR gene body methylation level. The overall survival analysis demonstrated that a high level of CFTR expression was associated with a worse survival rate in patients with JMML (P=0.039).
Conclusions: CFTR promoter hypermethylation may be a novel biomarker for the diagnosis, monitoring of disease progression, and prognosis of JMML.
Keywords: Bioinformatics analysis; cystic fibrosis transmembrane conductance regulator (CFTR); monozygotic twins.
2022 Translational Pediatrics. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-381/coif). The authors have no conflicts of interest to declare.
Figures





Similar articles
-
Integrated molecular profiling of juvenile myelomonocytic leukemia.Blood. 2018 Apr 5;131(14):1576-1586. doi: 10.1182/blood-2017-07-798157. Epub 2018 Feb 2. Blood. 2018. PMID: 29437595
-
[Relationship of Cystic Fibrosis Transmembrane Conductance Regulator Expression with Clinical Features and Prognosis in Patients with Acute Leukemia].Sichuan Da Xue Xue Bao Yi Xue Ban. 2019 May;50(3):420-424. Sichuan Da Xue Xue Bao Yi Xue Ban. 2019. PMID: 31631611 Chinese.
-
CREBBP is a target of epigenetic, but not genetic, modification in juvenile myelomonocytic leukemia.Clin Epigenetics. 2016 May 5;8:50. doi: 10.1186/s13148-016-0216-3. eCollection 2016. Clin Epigenetics. 2016. PMID: 27158276 Free PMC article.
-
Juvenile myelomonocytic leukemia-A comprehensive review and recent advances in management.Am J Blood Res. 2021 Feb 15;11(1):1-21. eCollection 2021. Am J Blood Res. 2021. PMID: 33796386 Free PMC article. Review.
-
Genomic and Epigenomic Landscape of Juvenile Myelomonocytic Leukemia.Cancers (Basel). 2022 Mar 4;14(5):1335. doi: 10.3390/cancers14051335. Cancers (Basel). 2022. PMID: 35267643 Free PMC article. Review.
References
-
- Hasle H, Kerndrup G, Jacobsen BB. Childhood myelodysplastic syndrome in Denmark: incidence and predisposing conditions. Leukemia 1995;9:1569-72. - PubMed
LinkOut - more resources
Full Text Sources
