Rapid detection of human coronavirus NL63 by isothermal reverse transcription recombinase polymerase amplification
- PMID: 36248766
- PMCID: PMC9546502
- DOI: 10.1016/j.jcvp.2022.100115
Rapid detection of human coronavirus NL63 by isothermal reverse transcription recombinase polymerase amplification
Abstract
Background: Human coronaviruses are one of the leading causes for respiratory tract infections and for frequent primary care consultation. The human coronavirus NL63 (HCoV..µNL63) is one representative of the seasonal coronaviruses and capable of infecting the upper and lower respiratory tract and causative agent for croup in children.
Objectives: For fast detection of HCoV-NL63, we developed an isothermal reverse transcription recombinase polymerase amplification (RT-RPA) assay.
Study design: The analytical sensitivities of the RT-RPA assay were identified for in vitro transcribed ribonucleic acid (RNA) and for genomic viral RNA from cell culture supernatant. Moreover, specificity was tested with nucleic acids from other human coronaviruses and a variety of clinically relevant respiratory viruses. Finally, a clinical nasopharyngeal swab sample with spiked genomic viral HCoV-NL63 RNA was analyzed.
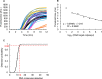
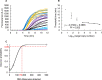
Results: Our HCoV-NL63 RT-RPA assay is highly specific and has an analytical sensitivity of 13 RNA molecules/reaction for in vitro transcribed RNA. For genomic viral RNA from cell culture supernatant spiked into a clinical nasopharyngeal swab sample the assay...s analytical sensitivity is 170 RNA molecules/reaction. The assay shows amplification of the lowest detectable target copy number after 8 minutes and 7 minutes, respectively.
Conclusions: We were able to design a sensitive and specific RT-RPA assay for the detection of HCoV-NL63. Additionally, the assay is characterized by short duration, isothermal amplification, and simple instrumentation.
Keywords: HCoV-NL63, human coronavirus NL63; LAMP, loop-mediated isothermal amplification; N-gene, nucleocapsid gene; RT-RPA, reverse transcription recombinase polymerase amplification; exo-IQ, internally quenched exo probe; human coronavirus NL63; recombinase polymerase amplification.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
[Visual Detection of Human Coronavirus NL63 by Reverse Transcription Loop-Mediated Isothermal Amplification].Bing Du Xue Bao. 2016 Jan;32(1):56-61. Bing Du Xue Bao. 2016. PMID: 27295884 Chinese.
-
"Rapid diagnosis of Cucumber mosaic virus in banana plants using a fluorescence-based real-time isothermal reverse transcription-recombinase polymerase amplification assay".J Virol Methods. 2019 Aug;270:52-58. doi: 10.1016/j.jviromet.2019.04.024. Epub 2019 Apr 29. J Virol Methods. 2019. PMID: 31047971
-
Rapid and sensitive detection of canine distemper virus by real-time reverse transcription recombinase polymerase amplification.BMC Vet Res. 2017 Aug 15;13(1):241. doi: 10.1186/s12917-017-1180-7. BMC Vet Res. 2017. PMID: 28810858 Free PMC article.
-
Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2.Emerg Microbes Infect. 2020 Dec;9(1):998-1007. doi: 10.1080/22221751.2020.1756698. Emerg Microbes Infect. 2020. PMID: 32306853 Free PMC article.
-
Advancements and applications of loop-mediated isothermal amplification technology: a comprehensive overview.Front Microbiol. 2024 Jul 17;15:1406632. doi: 10.3389/fmicb.2024.1406632. eCollection 2024. Front Microbiol. 2024. PMID: 39091309 Free PMC article. Review.
References
-
- Anthony R. Fehr, Stanley Perlman. Coronaviruses: an overview of their replication and pathogenesis, SpringerLink [Internet]. 2015 [cited 2020 Oct 18]. Available from: https://link.springer.com/protocol/10.1007/978-1-4939-2438-7_1 - DOI - PMC - PubMed
-
- Sánchez Choez X, Loaiza Martínez M, Vaca Tatamuez V, López Peña M, Manzano Pasquel A, Jimbo Sotomayor R. Medical cost of upper respiratory tract infections in children in ambulatory care. Value Health Reg. Issues. 2021;26:1–9. - PubMed
-
- Weston S, Frieman MB. Respiratory viruses. Encycl. Microbiol. 2019:85–101.
LinkOut - more resources
Full Text Sources
