Persistent but dysfunctional mucosal SARS-CoV-2-specific IgA and low lung IL-1β associate with COVID-19 fatal outcome: A cross-sectional analysis
- PMID: 36248831
- PMCID: PMC9560774
- DOI: 10.3389/fimmu.2022.842468
Persistent but dysfunctional mucosal SARS-CoV-2-specific IgA and low lung IL-1β associate with COVID-19 fatal outcome: A cross-sectional analysis
Abstract
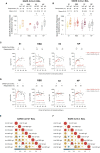
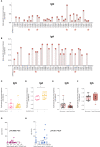
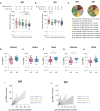
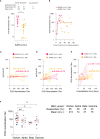
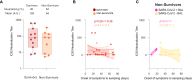
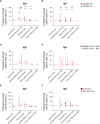
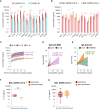
The role of the mucosal pulmonary antibody response in coronavirus disease 2019 (COVID-19) outcome remains unclear. Here, we found that in bronchoalveolar lavage (BAL) samples from 48 patients with severe COVID-19-infected with the ancestral Wuhan virus, mucosal IgG and IgA specific for S1, receptor-binding domain (RBD), S2, and nucleocapsid protein (NP) emerged in BAL containing viruses early in infection and persist after virus elimination, with more IgA than IgG for all antigens tested. Furthermore, spike-IgA and spike-IgG immune complexes were detected in BAL, especially when the lung virus has been cleared. BAL IgG and IgA recognized the four main RBD variants. BAL neutralizing titers were higher early in COVID-19 when virus replicates in the lung than later in infection after viral clearance. Patients with fatal COVID-19, in contrast to survivors, developed higher levels of mucosal spike-specific IgA than IgG but lost neutralizing activities over time and had reduced IL-1β in the lung. Altogether, mucosal spike and NP-specific IgG and S1-specific IgA persisting after lung severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) clearance and low pulmonary IL-1β correlate with COVID-19 fatal outcome. Thus, mucosal SARS-CoV-2-specific antibodies may have adverse functions in addition to protective neutralization.
Highlights: Mucosal pulmonary antibody response in COVID-19 outcome remains unclear. We show that in severe COVID-19 patients, mucosal pulmonary non-neutralizing SARS-CoV-2 IgA persit after viral clearance in the lung. Furthermore, low lung IL-1β correlate with fatal COVID-19. Altogether, mucosal IgA may exert harmful functions beside protective neutralization.
Keywords: COVID-19; IgA; SARS-CoV-2; inflammatory cytokine; mucosal immunity; severe infection.
Copyright © 2022 Ruiz, Siracusano, Cottignies-Calamarte, Tudor, Real, Zhu, Pastori, Capron, Rosenberg, Temperton, Cantoni, Liao, Ternette, Moine, Godement, Geri, Chiche, Annane, Cramer Bordé, Lopalco and Bomsel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
A Possible Protective Effect of IgA Against Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) in Bronchoalveolar Lavage in COVID-19 Patients Admitted to Intensive Care Unit.Viruses. 2024 Nov 28;16(12):1851. doi: 10.3390/v16121851. Viruses. 2024. PMID: 39772161 Free PMC article.
-
Distinct anti-NP, anti-RBD and anti-Spike antibody profiles discriminate death from survival in COVID-19.Front Immunol. 2023 Oct 9;14:1206979. doi: 10.3389/fimmu.2023.1206979. eCollection 2023. Front Immunol. 2023. PMID: 37876932 Free PMC article.
-
Assessment of S1-, S2-, and NCP-Specific IgM, IgA, and IgG Antibody Kinetics in Acute SARS-CoV-2 Infection by a Microarray and Twelve Other Immunoassays.J Clin Microbiol. 2021 Apr 20;59(5):e02890-20. doi: 10.1128/JCM.02890-20. Print 2021 Apr 20. J Clin Microbiol. 2021. PMID: 33602698 Free PMC article.
-
Therapeutic antibodies for COVID-19: is a new age of IgM, IgA and bispecific antibodies coming?MAbs. 2022 Jan-Dec;14(1):2031483. doi: 10.1080/19420862.2022.2031483. MAbs. 2022. PMID: 35220888 Free PMC article. Review.
-
Immunoglobulin A response to SARS-CoV-2 infection and immunity.Heliyon. 2024 Jan 3;10(1):e24031. doi: 10.1016/j.heliyon.2024.e24031. eCollection 2024 Jan 15. Heliyon. 2024. PMID: 38230244 Free PMC article. Review.
Cited by
-
Assessment of antibodies in the upper and lower human respiratory tract at steady state and after respiratory viral infection.Clin Transl Immunology. 2023 Aug 8;12(8):e1460. doi: 10.1002/cti2.1460. eCollection 2023. Clin Transl Immunology. 2023. PMID: 37564999 Free PMC article.
-
CGRP inhibits SARS-CoV-2 infection of bronchial epithelial cells, and its pulmonary levels correlate with viral clearance in critical COVID-19 patients.J Virol. 2024 Sep 17;98(9):e0012824. doi: 10.1128/jvi.00128-24. Epub 2024 Aug 20. J Virol. 2024. PMID: 39162434 Free PMC article.
-
The longitudinal characterization of immune responses in COVID-19 patients reveals novel prognostic signatures for disease severity, patients' survival and long COVID.Front Immunol. 2024 Jul 29;15:1381091. doi: 10.3389/fimmu.2024.1381091. eCollection 2024. Front Immunol. 2024. PMID: 39136010 Free PMC article.
-
A nasal vaccine with inactivated whole-virion elicits protective mucosal immunity against SARS-CoV-2 in mice.Front Immunol. 2023 Aug 31;14:1224634. doi: 10.3389/fimmu.2023.1224634. eCollection 2023. Front Immunol. 2023. PMID: 37720231 Free PMC article.
-
A Possible Protective Effect of IgA Against Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) in Bronchoalveolar Lavage in COVID-19 Patients Admitted to Intensive Care Unit.Viruses. 2024 Nov 28;16(12):1851. doi: 10.3390/v16121851. Viruses. 2024. PMID: 39772161 Free PMC article.
References
-
- Wu F, Wang Aojie , Liu Mei , Wang Qimin , Chen J, Xia S, et al. . Neutralizing antibody responses to sars-Cov-2 in a covid-19 recovered patient cohort and their implications. MedRxiv (2020). doi: 10.1101/2020.03.30.20047365 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

