Pathogenic variant c.1052T>A (p.Leu351Gln) in adenosine deaminase 2 impairs secretion and elevates type I IFN responsive gene expression
- PMID: 36248868
- PMCID: PMC9562767
- DOI: 10.3389/fimmu.2022.995191
Pathogenic variant c.1052T>A (p.Leu351Gln) in adenosine deaminase 2 impairs secretion and elevates type I IFN responsive gene expression
Abstract
Background: Adenosine deaminase 2 (ADA2) is a homodimeric, extracellular enzyme and putative growth factor that is produced by cells of the myeloid lineage and, catalytically, deaminates extracellular adenosine to inosine. Loss-of-(catalytic)-function variants in the ADA2 gene are associated with Deficiency of ADA2 (DADA2), an autosomal recessive disease associated with an unusually broad range of inflammatory manifestations including vasculitis, hematological defects and cytopenia. Previous work by our group led to the identification of ADA2 variants of novel association with DADA2, among which was a unique c.1052T>A (p.Leu351Gln; herein referred to as L351Q) variant located in the catalytic domain of the protein.
Methods: Mammalian (Flp-IN CHO) cells were engineered to stably express wild-type ADA2 and ADA2 protein variants, including the pathogenic L351Q variant identified in DADA2 patients. An enzyme assay and immunoblotting were used to assess ADA2 catalytic activity and secretion, respectively, and the outcome of experimentally induced inhibition of protein processing (Golgi transport and N-linked glycosylation) was assessed. Reverse transcription quantitative real-time PCR (RT-qPCR) was applied to determine the relative expression of Type I Interferon stimulated genes (ISGs), IFIT3 and IRF7.
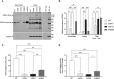
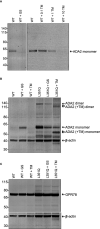
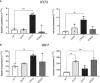
Results: In addition to abrogating catalytic activity, the L351Q variant impaired secretion of L351Q ADA2 resulting in an intracellular accumulation of L351Q ADA2 protein that was not observed in cells expressing wild-type ADA2 or other ADA2 protein variants. Retention of L351Q ADA2 was not attributable to impaired glycosylation on neighboring asparagine residues and did not impact cell growth or integrity. Constitutive expression of Type I ISGs IFIT3 and IRF7 was observed in cells expressing L351Q ADA2.
Conclusions: The impaired secretion of L351Q ADA2 may be an important factor leading to the severe phenotype observed in patients with this variant further emphasizing the importance of assessing impacts beyond catalytic activity when evaluating genotype-phenotype relationships in DADA2.
Keywords: adenosine deaminase 2 (ADA2); inflammation; pediatric; systemic vasculitis; type I Interferon (IFN).
Copyright © 2022 Bowers, Sundqvist, Dancey, Cabral and Brown.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
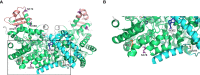
Similar articles
-
A Monogenic Disease with a Variety of Phenotypes: Deficiency of Adenosine Deaminase 2.J Rheumatol. 2020 Jan;47(1):117-125. doi: 10.3899/jrheum.181384. Epub 2019 May 1. J Rheumatol. 2020. PMID: 31043544
-
Comparison of disease phenotypes and mechanistic insight on causal variants in patients with DADA2.J Allergy Clin Immunol. 2023 Sep;152(3):771-782. doi: 10.1016/j.jaci.2023.04.014. Epub 2023 May 5. J Allergy Clin Immunol. 2023. PMID: 37150360
-
Identification of Novel Adenosine Deaminase 2 Gene Variants and Varied Clinical Phenotype in Pediatric Vasculitis.Arthritis Rheumatol. 2019 Oct;71(10):1747-1755. doi: 10.1002/art.40913. Epub 2019 Aug 26. Arthritis Rheumatol. 2019. PMID: 31008556
-
[ADA2, an Adenosine Deaminase Isozyme Acting as a Regulator of Autoinflammation].Yakugaku Zasshi. 2025;145(5):379-386. doi: 10.1248/yakushi.24-00194. Yakugaku Zasshi. 2025. PMID: 40307021 Review. Japanese.
-
Deficiency of Adenosine Deaminase 2 (DADA2): Updates on the Phenotype, Genetics, Pathogenesis, and Treatment.J Clin Immunol. 2018 Jul;38(5):569-578. doi: 10.1007/s10875-018-0525-8. Epub 2018 Jun 27. J Clin Immunol. 2018. PMID: 29951947 Free PMC article. Review.
Cited by
-
Getting to know adenosine deaminase 2 deficiency inside and out.J Allergy Clin Immunol. 2025 May;155(5):1451-1463. doi: 10.1016/j.jaci.2025.01.040. Epub 2025 Feb 14. J Allergy Clin Immunol. 2025. PMID: 39956283 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

