Crosstalk of necroptosis and pyroptosis defines tumor microenvironment characterization and predicts prognosis in clear cell renal carcinoma
- PMID: 36248876
- PMCID: PMC9561249
- DOI: 10.3389/fimmu.2022.1021935
Crosstalk of necroptosis and pyroptosis defines tumor microenvironment characterization and predicts prognosis in clear cell renal carcinoma
Abstract
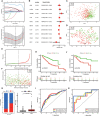
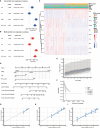
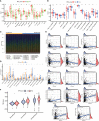
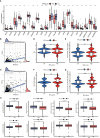
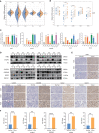
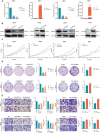
Pyroptosis and necroptosis are two recently identified forms of immunogenic cell death in the tumor microenvironment (TME), indicating a crucial involvement in tumor metastasis. However, the characteristics of necroptosis and pyroptosis that define tumor microenvironment and prognosis in ccRCC patients remain unknown. We systematically investigated the transcriptional variation and expression patterns of Necroptosis and Pyroptosis related genes (NPRGs). After screening the necroptosis-pyroptosis clusters, the potential functional annotation for clusters was explored by GSVA enrichment analysis. The Necroptosis-Pyroptosis Genes (NPG) scores were used for the prognosis model construction and validation. Then, the correlations of NPG score with clinical features, cancer stem cell (CSC) index, tumor mutation burden (TMB), TME, and Immune Checkpoint Genes (ICGs) were also individually explored to evaluate the prognosis predictive values in ccRCC. Microarray screenings identified 27 upregulated and 1 downregulated NPRGs. Ten overall survival associated NPRGs were filtered to construct the NPG prognostic model indicating a better prognostic signature for ccRCC patients with lower NPG scores (P< 0.001), which was verified using the external cohort. Univariate and multivariate analyses along with Kaplan-Meier survival analysis demonstrated that NPG score prognostic model could be applied as an independent prognostic factor, and AUC values of nomogram from 1- to 5- year overall survival with good agreement in calibration plots suggested that the proposed prognostic signature possessed good predictive capabilities in ccRCC. A high-/sNPG score is proven to be connected with tumor growth and immune-related biological processes, according to enriched GO, KEGG, and GSEA analyses. Comparing patients with a high-NPG score to those with a low-NPG score revealed significant differences in clinical characteristics, growth and recurrence of malignancies (CSC index), TME cell infiltration, and immunotherapeutic response (P< 0.005), potentially making the NPG score multifunctional in the clinical therapeutic setting. Furthermore, AIM2, CASP4, GSDMB, NOD2, and RBCK1 were also found to be highly expressed in ccRCC cell lines and tumor tissues, and GASP4 and GSDMB promote ccRCC cells' proliferation, migration, and invasion. This study firstly suggests that targeting the NPG score feature for TME characterization may lend novel insights into its clinical applications in the prognostic prediction of ccRCC.
Keywords: clear cell renal cell carcinoma; necroptosis; prognosis; pyroptosis; tumor microenvironment.
Copyright © 2022 Fu, Bao, Li, Li, Lin, Zhou, Li, Yan, Langston, Sun, Guo, Zhou, Chen, Liu, Zhao, Lu, Huang, Chen, Chung and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer HL declared a shared affiliation with the authors JL, ML and BC to the handling editor at the time of review.
Figures




Similar articles
-
Identification and validation of prognostic and tumor microenvironment characteristics of necroptosis index and BIRC3 in clear cell renal cell carcinoma.PeerJ. 2023 Dec 18;11:e16643. doi: 10.7717/peerj.16643. eCollection 2023. PeerJ. 2023. PMID: 38130918 Free PMC article.
-
A novel necroptosis-related long noncoding RNA model for predicting clinical features, immune characteristics, and therapeutic response in clear cell renal cell carcinoma.Front Immunol. 2023 Aug 2;14:1230267. doi: 10.3389/fimmu.2023.1230267. eCollection 2023. Front Immunol. 2023. PMID: 37600792 Free PMC article.
-
A novel prognostic signature for clear cell renal cell carcinoma constructed using necroptosis-related miRNAs.BMC Genomics. 2023 Mar 29;24(1):162. doi: 10.1186/s12864-023-09258-9. BMC Genomics. 2023. PMID: 36991314 Free PMC article.
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
A novel necroptosis-related gene index for predicting prognosis and a cold tumor immune microenvironment in stomach adenocarcinoma.Front Immunol. 2022 Oct 27;13:968165. doi: 10.3389/fimmu.2022.968165. eCollection 2022. Front Immunol. 2022. PMID: 36389725 Free PMC article. Review.
Cited by
-
Development and verification of a novel risk model related to ubiquitination linked with prognosis and therapeutic response in clear cell renal cell carcinoma.Sci Rep. 2024 Oct 27;14(1):25651. doi: 10.1038/s41598-024-75948-3. Sci Rep. 2024. PMID: 39463392 Free PMC article.
-
The prognostic and immunological role of MCM3 in pan-cancer and validation of prognosis in a clinical lower-grade glioma cohort.Front Pharmacol. 2024 Apr 18;15:1390615. doi: 10.3389/fphar.2024.1390615. eCollection 2024. Front Pharmacol. 2024. PMID: 38698811 Free PMC article.
-
Deciphering the tumour microenvironment of clear cell renal cell carcinoma: Prognostic insights from programmed death genes using machine learning.J Cell Mol Med. 2024 Jul;28(13):e18524. doi: 10.1111/jcmm.18524. J Cell Mol Med. 2024. PMID: 39011666 Free PMC article.
-
A novel cancer-associated fibroblast signature for kidney renal clear cell carcinoma via integrated analysis of single-cell and bulk RNA-sequencing.Discov Oncol. 2024 Jul 26;15(1):309. doi: 10.1007/s12672-024-01175-x. Discov Oncol. 2024. PMID: 39060620 Free PMC article.
-
Comprehensive analysis of LD-related genes signature for predicting prognosis and immunotherapy response in clear cell renal cell carcinoma.BMC Nephrol. 2024 Sep 10;25(1):298. doi: 10.1186/s12882-024-03735-3. BMC Nephrol. 2024. PMID: 39256647 Free PMC article.
References
-
- Key statistics about kidney cancer . Available at: https://www.cancer.org/cancer/kidney-cancer/about/key-statistics.html.
-
- Kidney cancer: Statistics . Available at: https://www.cancer.net/cancer-types/kidney-cancer/statistics#:~:text=Thi....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

