Epstein-Barr virus perpetuates B cell germinal center dynamics and generation of autoimmune-associated phenotypes in vitro
- PMID: 36248899
- PMCID: PMC9554744
- DOI: 10.3389/fimmu.2022.1001145
Epstein-Barr virus perpetuates B cell germinal center dynamics and generation of autoimmune-associated phenotypes in vitro
Abstract
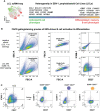
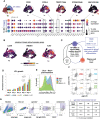
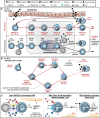
Human B cells encompass functionally diverse lineages and phenotypic states that contribute to protective as well as pathogenic responses. Epstein-Barr virus (EBV) provides a unique lens for studying heterogeneous B cell responses, given its adaptation to manipulate intrinsic cell programming. EBV promotes the activation, proliferation, and eventual outgrowth of host B cells as immortalized lymphoblastoid cell lines (LCLs) in vitro, which provide a foundational model of viral latency and lymphomagenesis. Although cellular responses and outcomes of infection can vary significantly within populations, investigations that capture genome-wide perspectives of this variation at single-cell resolution are in nascent stages. We have recently used single-cell approaches to identify EBV-mediated B cell heterogeneity in de novo infection and within LCLs, underscoring the dynamic and complex qualities of latent infection rather than a singular, static infection state. Here, we expand upon these findings with functional characterizations of EBV-induced dynamic phenotypes that mimic B cell immune responses. We found that distinct subpopulations isolated from LCLs could completely reconstitute the full phenotypic spectrum of their parental lines. In conjunction with conserved patterns of cell state diversity identified within scRNA-seq data, these data support a model in which EBV continuously drives recurrent B cell entry, progression through, and egress from the Germinal Center (GC) reaction. This "perpetual GC" also generates tangent cell fate trajectories including terminal plasmablast differentiation, which constitutes a replicative cul-de-sac for EBV from which lytic reactivation provides escape. Furthermore, we found that both established EBV latency and de novo infection support the development of cells with features of atypical memory B cells, which have been broadly associated with autoimmune disorders. Treatment of LCLs with TLR7 agonist or IL-21 was sufficient to generate an increased frequency of IgD-/CD27-/CD23-/CD38+/CD138+ plasmablasts. Separately, de novo EBV infection led to the development of CXCR3+/CD11c+/FCRL4+ B cells within days, providing evidence for possible T cell-independent origins of a recently described EBV-associated neuroinvasive CXCR3+ B cell subset in patients with multiple sclerosis. Collectively, this work reveals unexpected virus-driven complexity across infected cell populations and highlights potential roles of EBV in mediating or priming foundational aspects of virus-associated immune cell dysfunction in disease.
Keywords: B cell; Epstein-Barr virus; atypical memory B cells; autoimmunity; chronic infection; germinal center; lymphoblastoid cells; single-cell.
Copyright © 2022 SoRelle, Reinoso-Vizcaino, Horn and Luftig.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases.Autoimmunity. 2008 May;41(4):298-328. doi: 10.1080/08916930802024772. Autoimmunity. 2008. PMID: 18432410 Review.
-
Increased expression of Toll-like receptors 7 and 9 in myasthenia gravis thymus characterized by active Epstein-Barr virus infection.Immunobiology. 2016 Apr;221(4):516-27. doi: 10.1016/j.imbio.2015.12.007. Epub 2015 Dec 12. Immunobiology. 2016. PMID: 26723518
-
Identification of Host Biomarkers of Epstein-Barr Virus Latency IIb and Latency III.mBio. 2019 Jul 2;10(4):e01006-19. doi: 10.1128/mBio.01006-19. mBio. 2019. PMID: 31266868 Free PMC article.
-
Time-resolved transcriptomes reveal diverse B cell fate trajectories in the early response to Epstein-Barr virus infection.Cell Rep. 2022 Aug 30;40(9):111286. doi: 10.1016/j.celrep.2022.111286. Cell Rep. 2022. PMID: 36044865 Free PMC article.
-
Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation.Adv Virus Res. 2014;88:279-313. doi: 10.1016/B978-0-12-800098-4.00006-4. Adv Virus Res. 2014. PMID: 24373315 Free PMC article. Review.
Cited by
-
The Role of CXCR3 in Nervous System-Related Diseases.Mediators Inflamm. 2024 Oct 11;2024:8347647. doi: 10.1155/2024/8347647. eCollection 2024. Mediators Inflamm. 2024. PMID: 39429695 Free PMC article. Review.
-
E2F1 suppresses Epstein-Barr virus lytic reactivation through cellular and viral transcriptional networks.PLoS Pathog. 2025 Aug 7;21(8):e1013410. doi: 10.1371/journal.ppat.1013410. eCollection 2025 Aug. PLoS Pathog. 2025. PMID: 40773523 Free PMC article.
-
Characterization of latently infected EBV+ antibody-secreting B cells isolated from ovarian tumors and malignant ascites.Front Immunol. 2024 Jul 17;15:1379175. doi: 10.3389/fimmu.2024.1379175. eCollection 2024. Front Immunol. 2024. PMID: 39086481 Free PMC article.
-
EBV induces CNS homing of B cells attracting inflammatory T cells.Nature. 2025 Aug 6. doi: 10.1038/s41586-025-09378-0. Online ahead of print. Nature. 2025. PMID: 40770101
-
Advances in Single-Cell Sequencing for Infectious Diseases: Progress and Perspectives.Adv Sci (Weinh). 2025 Aug;12(32):e15678. doi: 10.1002/advs.202415678. Epub 2025 Jul 4. Adv Sci (Weinh). 2025. PMID: 40613694 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

