RNA polymerase II-driven CRISPR-Cas9 system for efficient non-growth-biased metabolic engineering of Kluyveromyces marxianus
- PMID: 36249306
- PMCID: PMC9558044
- DOI: 10.1016/j.mec.2022.e00208
RNA polymerase II-driven CRISPR-Cas9 system for efficient non-growth-biased metabolic engineering of Kluyveromyces marxianus
Abstract
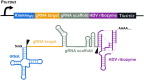
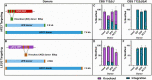
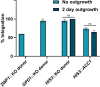
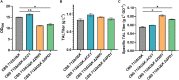
The thermotolerant yeast Kluyveromyces marxianus has gained significant attention in recent years as a promising microbial candidate for industrial biomanufacturing. Despite several contributions to the expanding molecular toolbox for gene expression and metabolic engineering of K. marxianus, there remains a need for a more efficient and versatile genome editing platform. To address this, we developed a CRISPR-based editing system that enables high efficiency marker-less gene disruptions and integrations using only 40 bp homology arms in NHEJ functional and non-functional K. marxianus strains. The use of a strong RNA polymerase II promoter allows efficient expression of gRNAs flanked by the self-cleaving RNA structures, tRNA and HDV ribozyme, from a single plasmid co-expressing a codon optimized Cas9. Implementing this system resulted in nearly 100% efficiency of gene disruptions in both NHEJ-functional and NHEJ-deficient K. marxianus strains, with donor integration efficiencies reaching 50% and 100% in the two strains, respectively. The high gRNA targeting performance also proved instrumental for selection of engineered strains with lower growth rate but improved polyketide biosynthesis by avoiding an extended outgrowth period, a common method used to enrich for edited cells but that fails to recover advantageous mutants with even slightly impaired fitness. Finally, we provide the first demonstration of simultaneous, markerless integrations at multiple loci in K. marxianus using a 2.6 kb and a 7.6 kb donor, achieving a dual integration efficiency of 25.5% in a NHEJ-deficient strain. These results highlight both the ease of use and general robustness of this system for rapid and flexible metabolic engineering in this non-conventional yeast.
Keywords: CRISPR-Cas9; Kluyveromyces marxianus; Metabolic engineering; Polyketides; Thermotolerant yeast.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
CRISPR-Cas9 mediated genome editing of Kluyveromyces marxianus for iterative, multiplexed gene disruption and pathway integration.Biotechnol Bioeng. 2024 Oct;121(10):3269-3282. doi: 10.1002/bit.28736. Epub 2024 May 22. Biotechnol Bioeng. 2024. PMID: 38778433
-
Engineering Kluyveromyces marxianus as a Robust Synthetic Biology Platform Host.mBio. 2018 Sep 25;9(5):e01410-18. doi: 10.1128/mBio.01410-18. mBio. 2018. PMID: 30254120 Free PMC article.
-
Development of a comprehensive set of tools for genome engineering in a cold- and thermo-tolerant Kluyveromyces marxianus yeast strain.Sci Rep. 2017 Aug 21;7(1):8993. doi: 10.1038/s41598-017-08356-5. Sci Rep. 2017. PMID: 28827530 Free PMC article.
-
Integration of comprehensive data and biotechnological tools for industrial applications of Kluyveromyces marxianus.Appl Microbiol Biotechnol. 2020 Jan;104(2):475-488. doi: 10.1007/s00253-019-10224-3. Epub 2019 Nov 28. Appl Microbiol Biotechnol. 2020. PMID: 31781815 Review.
-
Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris.J Biotechnol. 2016 Oct 10;235:139-49. doi: 10.1016/j.jbiotec.2016.03.027. Epub 2016 Mar 22. J Biotechnol. 2016. PMID: 27015975 Review.
Cited by
-
Engineering a carbon source-responsive promoter for improved biosynthesis in the non-conventional yeast Kluyveromyces marxianus.Metab Eng Commun. 2024 May 20;18:e00238. doi: 10.1016/j.mec.2024.e00238. eCollection 2024 Jun. Metab Eng Commun. 2024. PMID: 38845682 Free PMC article.
-
Metabolic engineering and adaptive laboratory evolution of Kluyveromyces Marxianus for lactic acid production.Microb Cell Fact. 2025 Aug 4;24(1):179. doi: 10.1186/s12934-025-02805-x. Microb Cell Fact. 2025. PMID: 40760443 Free PMC article.
-
Gene disruption via a transient hypercompact CRISPR-AsCas12f1 system in Kluyveromyces marxianus.Biotechnol Lett. 2025 Jul 31;47(4):84. doi: 10.1007/s10529-025-03626-z. Biotechnol Lett. 2025. PMID: 40742466
References
-
- Amrane A., Prigent Y. Effect of culture conditions of Kluyveromyces marxianus on its autolysis, and process optimization. Bioprocess Eng. 1998;18:383–388. doi: 10.1007/pl00008998. - DOI

