Examining the diversity of structural motifs in fungal glycome
- PMID: 36249563
- PMCID: PMC9535381
- DOI: 10.1016/j.csbj.2022.09.040
Examining the diversity of structural motifs in fungal glycome
Abstract
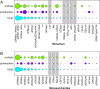
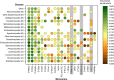
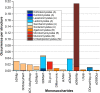
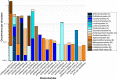
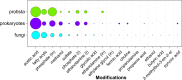
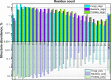
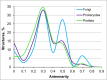
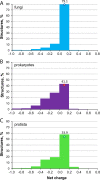
In this paper, we present the results of a systematic statistical analysis of the fungal glycome in comparison with the prokaryotic and protistal glycomes as described in the scientific literature and presented in the Carbohydrate Structure Database (CSDB). The monomeric and dimeric compositions of glycans, their non-carbohydrate modifications, glycosidic linkages, sizes of structures, branching degree and net charge are assessed. The obtained information can help elucidating carbohydrate molecular markers for various fungal classes which, in its turn, can be demanded for the development of diagnostic tools and carbohydrate-based vaccines against pathogenic fungi. It can also be useful for revealing specific glycosyltransferases active in a particular fungal species.
Keywords: Bacteria; CSDB; Carbohydrate; Diversity; Fungi; Glycan; Glycome; Protista.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Newbound M., McCarthy M.A., Lebel T. Landscape Urban Plann. 2010;96:138.
-
- Latgé J.-P., Beauvais A. Curr Opin Microbiol. 2014;20:111. - PubMed
LinkOut - more resources
Full Text Sources
