Regions with two amino acids in protein sequences: A step forward from homorepeats into the low complexity landscape
- PMID: 36249567
- PMCID: PMC9550522
- DOI: 10.1016/j.csbj.2022.09.011
Regions with two amino acids in protein sequences: A step forward from homorepeats into the low complexity landscape
Abstract
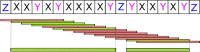
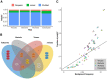
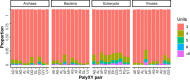
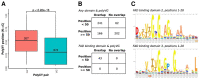
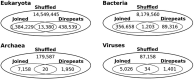
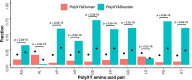
Low complexity regions (LCRs) differ in amino acid composition from the background provided by the corresponding proteomes. The simplest LCRs are homorepeats (or polyX), regions composed of mostly-one amino acid type. Extensive research has been done to characterize homorepeats, and their taxonomic, functional and structural features depend on the amino acid type and sequence context. From them, the next step towards the study of LCRs are the regions composed of two types of amino acids, which we call polyXY. We classify polyXY in three categories based on the arrangement of the two amino acid types 'X' and 'Y': direpeats (e.g. 'XYXYXY'), joined (e.g. 'XXXYYY') and shuffled (e.g. 'XYYXXY'). We developed a script to search for polyXY, and located them in a comprehensive set of 20,340 reference proteomes. These results are available in a dedicated web server called XYs, in which the user can also submit their own protein datasets to detect polyXY. We studied the distribution of polyXY types by amino acid pair XY and category, and show that polyXY in Eukaryota are mainly located within intrinsically disordered regions. Our study provides a first step towards the characterization of polyXY as protein motifs.
Keywords: Linear motifs; Low complexity regions; Protein sequence analysis; polyXY.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
The nucleotide landscape of polyXY regions.Comput Struct Biotechnol J. 2023 Oct 31;21:5408-5412. doi: 10.1016/j.csbj.2023.10.054. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38022702 Free PMC article.
-
One Step Closer to the Understanding of the Relationship IDR-LCR-Structure.Genes (Basel). 2023 Aug 28;14(9):1711. doi: 10.3390/genes14091711. Genes (Basel). 2023. PMID: 37761851 Free PMC article.
-
Low Complexity Induces Structure in Protein Regions Predicted as Intrinsically Disordered.Biomolecules. 2022 Aug 10;12(8):1098. doi: 10.3390/biom12081098. Biomolecules. 2022. PMID: 36008992 Free PMC article.
-
Structure-function relationships in protein homorepeats.Curr Opin Struct Biol. 2023 Dec;83:102726. doi: 10.1016/j.sbi.2023.102726. Epub 2023 Nov 2. Curr Opin Struct Biol. 2023. PMID: 37924569 Review.
-
Amino acid homorepeats in proteins.Nat Rev Chem. 2020 Aug;4(8):420-434. doi: 10.1038/s41570-020-0204-1. Epub 2020 Jul 21. Nat Rev Chem. 2020. PMID: 37127972 Review.
Cited by
-
The nucleotide landscape of polyXY regions.Comput Struct Biotechnol J. 2023 Oct 31;21:5408-5412. doi: 10.1016/j.csbj.2023.10.054. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38022702 Free PMC article.
-
One Step Closer to the Understanding of the Relationship IDR-LCR-Structure.Genes (Basel). 2023 Aug 28;14(9):1711. doi: 10.3390/genes14091711. Genes (Basel). 2023. PMID: 37761851 Free PMC article.
-
Identification of Low-Complexity Domains by Compositional Signatures Reveals Class-Specific Frequencies and Functions Across the Domains of Life.PLoS Comput Biol. 2024 May 15;20(5):e1011372. doi: 10.1371/journal.pcbi.1011372. eCollection 2024 May. PLoS Comput Biol. 2024. PMID: 38748749 Free PMC article.
-
Phase separating Rho: a widespread regulatory function of disordered regions in proteins revealed in bacteria.Signal Transduct Target Ther. 2023 Jun 21;8(1):253. doi: 10.1038/s41392-023-01505-5. Signal Transduct Target Ther. 2023. PMID: 37344523 Free PMC article. No abstract available.
References
-
- Mier P., Alanis-Lobato G., Andrade-Navarro M.A. Context characterization of amino acid homorepeats using evolution, position, and order. Proteins. 2017;85(4):709–719. - PubMed
-
- Romov P.A., Li F., Lipke P.N., Epstein S.L., Qiu W.-G. Comparative genomics reveals long, evolutionarily conserved, low-complexity islands in yeast proteins. J Mol Biol. 2006;63(3):415–425. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
