Improved Uveal Melanoma Copy Number Subtypes Including an Ultra-High-Risk Group
- PMID: 36249692
- PMCID: PMC9559896
- DOI: 10.1016/j.xops.2022.100121
Improved Uveal Melanoma Copy Number Subtypes Including an Ultra-High-Risk Group
Abstract
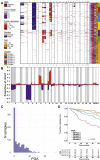
Purpose: To evaluate the clinical relevance of low-frequency copy number aberrations (CNAs) in uveal melanoma (UM) and to discern residual genomic and clinical heterogeneity within established molecular subtypes based on genome-wide CNA profiling of 921 primary tumors.
Design: Retrospective single-center case series.
Participants: Patients with primary UM referred for genetic testing between 2008 and 2016 (n = 921). The Cancer Genome Atlas cohort with clinical outcome data available (n = 70) was used to validate findings.
Methods: Genome-wide CNAs were generated for primary tumors from 921 patients and for 19 metastatic UM (mUM) in the liver. Of the 921 patients, metastatic outcome was known for 678 patients with a median time to metastasis of 4.5 years. The primary tumors were processed on the Affymetrix arrays SNP-5.0 (n = 140), SNP-6.0 (n = 359), or CytoScanHD (n = 422), and the metastatic tumors on the CytoScanHD array (n = 19). Recurrent CNAs were identified, and the prognostic effect of individual CNAs and multiple CNA clustering strategies, including more specific molecular subgroups with rare CNAs, were evaluated.
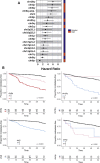
Main outcome measures: CNA recurrence, and effect of CNAs and derived molecular subtypes on metastatic-free survival.
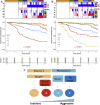
Results: Genomic profiling revealed CNAs associated with risk of metastasis and demonstrated a strong association between chromosomal instability and patient prognosis. Using standard prognostic CNAs, 6 clusters were detected, and inclusion of chromosome 16q deletion revealed an additional cluster. Of these 7 genomic clusters, 5 patient groups showed distinct rates of metastasis, indicating that different genomic patterns can have similar patient outcomes. A small group of patients with a significantly higher rate of metastasis was characterized by monosomy 3, 8q amplification, and deletion of 1p or 16q. Although this ultra-high-risk group accounts for only 7% of this cohort, 88% demonstrated metastasis within 4 years, compared with 45% in the second-highest risk group.
Conclusions: These results suggest that 1p and 16q deletion should be incorporated in clinical assays to assess prognosis at diagnosis and to guide enrollment in clinical trials for adjuvant therapies.
Keywords: 16q deletion; CNA, copy number aberration; Copy number profile; FNAB, fine-needle aspiration biopsy; HR, hazard ratio; Molecular risk groups; Prognosis; TCGA, The Cancer Genome Atlas; UM, uveal melanoma; Uveal melanoma; mUM, metastatic uveal melanoma.
© 2022 by the American Academy of Ophthalmology.
Figures


References
-
- Kujala E., Mäkitie T., Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44:4651–4659. - PubMed
-
- Kath R., Hayungs J., Bornfeld N., et al. Prognosis and treatment of disseminated uveal melanoma. Cancer. 1993;72:2219–2223. - PubMed
-
- Shields C.L., Ganguly A., Bianciotto C.G., et al. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2011;118:396–401. - PubMed
LinkOut - more resources
Full Text Sources

