GPI-anchored glutathione S-transferase as marker allows affinity sorting of transfection-positive cells
- PMID: 36250010
- PMCID: PMC9558730
- DOI: 10.3389/fmolb.2022.1016090
GPI-anchored glutathione S-transferase as marker allows affinity sorting of transfection-positive cells
Abstract
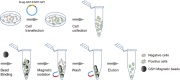
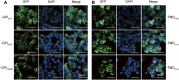
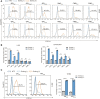
Cell transfection efficiency is still a limiting factor in gene function research. A method that allows isolation and enrichment of the transfection-positive cells is an effective solution. Here, we report a transfection-positive cell sorting system that utilizes GPI-anchored GST (Glutathione S-transferase) as a plasmid marker. The Glutathione S-transferase fusion protein will be expressed and displayed on the cell surface through GPI anchor, and hence permits the positive cells to be isolated using Glutathione (GSH) Magnetic Beads. We prove that the system works efficiently in both the adherent Lenti-X 293T cells and the suspension K-562 cells. The affinity cell sorting procedure efficiently enriched positive cells from 20% to 98% in K-562 cells. The applications in gene knockdown and overexpression experiments in K-562 cells dramatically enhanced the extent of gene alteration, with the gene knockdown efficiency increasing from 7% to 60% and the gene overexpression level rising from 47 to 253 times. This Glutathione S-transferase affinity transfection-positive cell sorting method is simple and fast to operate, large-instrument free, low cost, and hence possesses great potential in gene function study in vitro.
Keywords: GPI; cell sorting; cell transfection; glutathione; glutathione S-transferase.
Copyright © 2022 Ma, Yang, Zuo and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

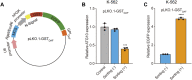
Similar articles
-
A Gene Transfer-Positive Cell Sorting System Utilizing Membrane-Anchoring Affinity Tag.Front Bioeng Biotechnol. 2022 Jun 16;10:930966. doi: 10.3389/fbioe.2022.930966. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35782508 Free PMC article.
-
Caveolin transfection results in caveolae formation but not apical sorting of glycosylphosphatidylinositol (GPI)-anchored proteins in epithelial cells.J Cell Biol. 1998 Feb 9;140(3):617-26. doi: 10.1083/jcb.140.3.617. J Cell Biol. 1998. PMID: 9456321 Free PMC article.
-
Glutathione-S-transferase-green fluorescent protein fusion protein reveals slow dissociation from high site density beads and measures free GSH.Cytometry A. 2006 May;69(5):326-34. doi: 10.1002/cyto.a.20259. Cytometry A. 2006. PMID: 16604533
-
Sorting of GPI-anchored proteins from yeast to mammals--common pathways at different sites?J Cell Sci. 2014 Jul 1;127(Pt 13):2793-801. doi: 10.1242/jcs.148056. Epub 2014 Jun 6. J Cell Sci. 2014. PMID: 24906797 Review.
-
Glutathione-S-transferase-fusion based assays for studying protein-protein interactions.Methods Mol Biol. 2004;261:175-86. doi: 10.1385/1-59259-762-9:175. Methods Mol Biol. 2004. PMID: 15064458 Review.
Cited by
-
Investigation of Glycosylphosphatidylinositol (GPI)-Plasma Membrane Interaction in Live Cells and the Influence of GPI Glycan Structure on the Interaction.Chemistry. 2024 Feb 7;30(8):e202303047. doi: 10.1002/chem.202303047. Epub 2023 Dec 14. Chemistry. 2024. PMID: 37966101 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

