Improvement of native structure-based peptides as efficient inhibitors of protein-protein interactions of SARS-CoV-2 spike protein and human ACE2
- PMID: 36250011
- PMCID: PMC9555309
- DOI: 10.3389/fmolb.2022.983014
Improvement of native structure-based peptides as efficient inhibitors of protein-protein interactions of SARS-CoV-2 spike protein and human ACE2
Abstract
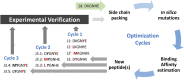
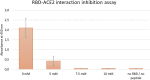
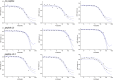
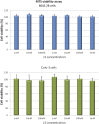
New pathogens responsible for novel human disease outbreaks in the last two decades are mainly the respiratory system viruses. Not different was the last pandemic episode, caused by infection of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). One of the extensively explored targets, in the recent scientific literature, as a possible way for rapid development of COVID-19 specific drug(s) is the interaction between the receptor-binding domain of the virus' spike (S) glycoprotein and human receptor angiotensin-converting enzyme 2 (hACE2). This protein-protein recognition process is involved in the early stages of the SARS-CoV-2 life cycle leading to the host cell membrane penetration. Thus, disrupting this interaction may block or significantly reduce the infection caused by the novel pathogen. Previously we have designed (by in silico structure-based analysis) three very short peptides having sequences inspirited by hACE2 native fragments, which effectively bind to the SARS-CoV-2 S protein and block its interaction with the human receptor. In continuation of the above mentioned studies, here we presented an application of molecular modeling approach resulting in improved binding affinity of the previously proposed ligand and its enhanced ability to inhibit meaningful host-virus protein-protein interaction. The new optimized hexapeptide binds to the virus protein with affinity one magnitude higher than the initial ligand and, as a very short peptide, has also great potential for further drug development. The peptide-based strategy is rapid and cost-effective for developing and optimizing efficient protein-protein interactions disruptors and may be successfully applied to discover antiviral candidates against other future emerging human viral infections.
Keywords: ACE2; COVID-19; SARS-CoV-2; angiotensin-converting enzyme-2; coronavirus; drug design; inhibitors of protein-protein interactions; peptides.
Copyright © 2022 Odolczyk, Klim, Podsiadła-Białoskórska, Winiewska-Szajewska, Szolajska, Zielenkiewicz, Poznański and Zielenkiewicz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
-
Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain.F1000Res. 2020 Jun 9;9:576. doi: 10.12688/f1000research.24074.1. eCollection 2020. F1000Res. 2020. PMID: 32802318 Free PMC article.
-
Pathway enrichment analysis of virus-host interactome and prioritization of novel compounds targeting the spike glycoprotein receptor binding domain-human angiotensin-converting enzyme 2 interface to combat SARS-CoV-2.J Biomol Struct Dyn. 2022 Apr;40(6):2701-2714. doi: 10.1080/07391102.2020.1841681. Epub 2020 Nov 4. J Biomol Struct Dyn. 2022. PMID: 33146070 Free PMC article.
-
De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2.Aging (Albany NY). 2020 Jun 16;12(12):11263-11276. doi: 10.18632/aging.103416. Epub 2020 Jun 16. Aging (Albany NY). 2020. PMID: 32544884 Free PMC article.
-
Exploring Spike Protein as Potential Target of Novel Coronavirus and to Inhibit the Viability Utilizing Natural Agents.Curr Drug Targets. 2021;22(17):2006-2020. doi: 10.2174/1389450122666210309105820. Curr Drug Targets. 2021. PMID: 33687893 Review.
Cited by
-
Peptide-Based Inhibitors of Protein-Protein Interactions (PPIs): A Case Study on the Interaction Between SARS-CoV-2 Spike Protein and Human Angiotensin-Converting Enzyme 2 (hACE2).Biomedicines. 2024 Oct 16;12(10):2361. doi: 10.3390/biomedicines12102361. Biomedicines. 2024. PMID: 39457672 Free PMC article. Review.
-
Study of Potential Blocking Peptides Targeting the SARS-CoV-2 RBD/hACE2 Interaction.Pharmaceuticals (Basel). 2024 Sep 20;17(9):1240. doi: 10.3390/ph17091240. Pharmaceuticals (Basel). 2024. PMID: 39338402 Free PMC article.
-
Editorial: Antiviral drug discovery against pathogens of pandemic concern: Advancements in target site identification and structure-based drug development.Front Mol Biosci. 2023 Mar 9;10:1165208. doi: 10.3389/fmolb.2023.1165208. eCollection 2023. Front Mol Biosci. 2023. PMID: 36968270 Free PMC article. No abstract available.
References
-
- Barh D., Tiwari S., Silva Andrade B., Giovanetti M., Almeida Costa E., Kumavath R., et al. (2020). Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain, 9, 576. 10.12688/f1000research.24074.1 Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain F1000Res - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

