Natural antisense transcripts as drug targets
- PMID: 36250017
- PMCID: PMC9563854
- DOI: 10.3389/fmolb.2022.978375
Natural antisense transcripts as drug targets
Abstract
The recent discovery of vast non-coding RNA-based regulatory networks that can be easily modulated by nucleic acid-based drugs has opened numerous new therapeutic possibilities. Long non-coding RNA, and natural antisense transcripts (NATs) in particular, play a significant role in networks that involve a wide variety of disease-relevant biological mechanisms such as transcription, splicing, translation, mRNA degradation and others. Currently, significant efforts are dedicated to harnessing these newly emerging NAT-mediated biological mechanisms for therapeutic purposes. This review will highlight the recent clinical and pre-clinical developments in this field and survey the advances in nucleic acid-based drug technologies that make these developments possible.
Keywords: anisense oligonucleotides; long nocoding RNA; natural antisense transcript (NAT); nucleic acid based therapeutics; posttranscriptional regulation.
Copyright © 2022 Khorkova, Stahl, Joji, Volmar, Zeier and Wahlestedt.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
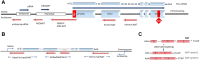
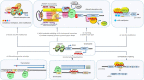

References
-
- Alterman J. F., Godinho B. M. D. C., Hassler M. R., Ferguson C. M., Echeverria D., Sapp E., et al. (2019). A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 37 (8), 884–894. 10.1038/s41587-019-0205-0 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

