Impaired TIGIT expression on B cells drives circulating follicular helper T cell expansion in multiple sclerosis
- PMID: 36250467
- PMCID: PMC9566906
- DOI: 10.1172/JCI156254
Impaired TIGIT expression on B cells drives circulating follicular helper T cell expansion in multiple sclerosis
Abstract
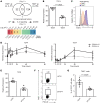
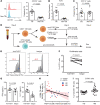
B cell depletion in patients with relapsing-remitting multiple sclerosis (RRMS) markedly prevents new MRI-detected lesions and disease activity, suggesting the hypothesis that altered B cell function leads to the activation of T cells driving disease pathogenesis. Here, we performed comprehensive analyses of CD40 ligand- (CD40L-) and IL-21-stimulated memory B cells from patients with MS and healthy age-matched controls, modeling the help of follicular helper T cells (Tfh cells), and found a differential gene expression signature in multiple B cell pathways. Most striking was the impaired TIGIT expression on MS-derived B cells mediated by dysregulation of the transcription factor TCF4. Activated circulating Tfh cells (cTfh cells) expressed CD155, the ligand of TIGIT, and TIGIT on B cells revealed their capacity to suppress the proliferation of IL-17-producing cTfh cells via the TIGIT/CD155 axis. Finally, CCR6+ cTfh cells were significantly increased in patients with MS, and their frequency was inversely correlated with that of TIGIT+ B cells. Together, these data suggest that the dysregulation of negative feedback loops between TIGIT+ memory B cells and cTfh cells in MS drives the activated immune system in this disease.
Keywords: Adaptive immunity; Autoimmunity; Immunology.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

