Cellular interferon-gamma and interleukin-2 responses to SARS-CoV-2 structural proteins are broader and higher in those vaccinated after SARS-CoV-2 infection compared to vaccinees without prior SARS-CoV-2 infection
- PMID: 36251675
- PMCID: PMC9576055
- DOI: 10.1371/journal.pone.0276241
Cellular interferon-gamma and interleukin-2 responses to SARS-CoV-2 structural proteins are broader and higher in those vaccinated after SARS-CoV-2 infection compared to vaccinees without prior SARS-CoV-2 infection
Abstract
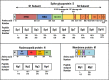
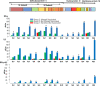
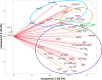
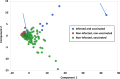
Class I- and Class II-restricted epitopes have been identified across the SARS-CoV-2 structural proteome. Vaccine-induced and post-infection SARS-CoV-2 T-cell responses are associated with COVID-19 recovery and protection, but the precise role of T-cell responses remains unclear, and how post-infection vaccination ('hybrid immunity') further augments this immunity To accomplish these goals, we studied healthy adult healthcare workers who were (a) uninfected and unvaccinated (n = 12), (b) uninfected and vaccinated with Pfizer-BioNTech BNT162b2 vaccine (2 doses n = 177, one dose n = 1) or Moderna mRNA-1273 vaccine (one dose, n = 1), and (c) previously infected with SARS-CoV-2 and vaccinated (BNT162b2, two doses, n = 6, one dose n = 1; mRNA-1273 two doses, n = 1). Infection status was determined by repeated PCR testing of participants. We used FluoroSpot Interferon-gamma (IFN-γ) and Interleukin-2 (IL-2) assays, using subpools of 15-mer peptides covering the S (10 subpools), N (4 subpools) and M (2 subpools) proteins. Responses were expressed as frequencies (percent positive responders) and magnitudes (spot forming cells/106 cytokine-producing peripheral blood mononuclear cells [PBMCs]). Almost all vaccinated participants with no prior infection exhibited IFN-γ, IL-2 and IFN-γ+IL2 responses to S glycoprotein subpools (89%, 93% and 27%, respectively) mainly directed to the S2 subunit and were more robust than responses to the N or M subpools. However, in previously infected and vaccinated participants IFN-γ, IL-2 and IFN-γ+IL2 responses to S subpools (100%, 100%, 88%) were substantially higher than vaccinated participants with no prior infection and were broader and directed against nine of the 10 S glycoprotein subpools spanning the S1 and S2 subunits, and all the N and M subpools. 50% of uninfected and unvaccinated individuals had IFN-γ but not IL2 or IFN-γ+IL2 responses against one S and one M subpools that were not increased after vaccination of uninfected or SARS-CoV-2-infected participants. Summed IFN-γ, IL-2, and IFN-γ+IL2 responses to S correlated with IgG responses to the S glycoprotein. These studies demonstrated that vaccinations with BNT162b2 or mRNA-1273 results in T cell-specific responses primarily against epitopes in the S2 subunit of the S glycoprotein, and that individuals that are vaccinated after SARS-CoV-2 infection develop broader and greater T cell responses to S1 and S2 subunits as well as the N and M proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
CHARM: COVID-19 Health Action Response for Marines-Association of antigen-specific interferon-gamma and IL2 responses with asymptomatic and symptomatic infections after a positive qPCR SARS-CoV-2 test.PLoS One. 2022 Apr 7;17(4):e0266691. doi: 10.1371/journal.pone.0266691. eCollection 2022. PLoS One. 2022. PMID: 35390102 Free PMC article.
-
T-cell and antibody responses to first BNT162b2 vaccine dose in previously infected and SARS-CoV-2-naive UK health-care workers: a multicentre prospective cohort study.Lancet Microbe. 2022 Jan;3(1):e21-e31. doi: 10.1016/S2666-5247(21)00275-5. Epub 2021 Nov 9. Lancet Microbe. 2022. PMID: 34778853 Free PMC article.
-
Cell immunity to SARS-CoV-2 after natural infection and/or different vaccination regimens.Front Cell Infect Microbiol. 2024 Mar 20;14:1370859. doi: 10.3389/fcimb.2024.1370859. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38572317 Free PMC article.
-
The temporal course of T- and B-cell responses to vaccination with BNT162b2 and mRNA-1273.Clin Microbiol Infect. 2022 May;28(5):701-709. doi: 10.1016/j.cmi.2021.09.006. Epub 2021 Sep 20. Clin Microbiol Infect. 2022. PMID: 34547457 Free PMC article.
-
Role of T cells in severe COVID-19 disease, protection, and long term immunity.Immunogenetics. 2023 Jun;75(3):295-307. doi: 10.1007/s00251-023-01294-9. Epub 2023 Feb 8. Immunogenetics. 2023. PMID: 36752852 Free PMC article. Review.
Cited by
-
The infectious diseases clinical research program acute respiratory infection repository protocol: Opportunities to understand current and future epidemics.PLoS One. 2025 Jul 23;20(7):e0317065. doi: 10.1371/journal.pone.0317065. eCollection 2025. PLoS One. 2025. PMID: 40700358 Free PMC article.
-
A randomized clinical trial of the impact of melatonin on influenza vaccine: Outcomes from the melatonin and vaccine response immunity and chronobiology study (MAVRICS).Hum Vaccin Immunother. 2024 Dec 31;20(1):2419742. doi: 10.1080/21645515.2024.2419742. Epub 2024 Nov 13. Hum Vaccin Immunother. 2024. PMID: 39539030 Free PMC article. Clinical Trial.
-
Development of a Candidate TMV Epitope Display Vaccine against SARS-CoV-2.Vaccines (Basel). 2024 Apr 23;12(5):448. doi: 10.3390/vaccines12050448. Vaccines (Basel). 2024. PMID: 38793699 Free PMC article.
-
Comparing population-level humoral and cellular immunity to SARS-Cov-2 in Bangalore, India.Sci Rep. 2024 Mar 8;14(1):5758. doi: 10.1038/s41598-024-54922-z. Sci Rep. 2024. PMID: 38459035 Free PMC article.
-
Distinct Omicron longitudinal memory T cell profile and T cell receptor repertoire associated with COVID-19 hospitalisation.Front Immunol. 2025 Apr 2;16:1549570. doi: 10.3389/fimmu.2025.1549570. eCollection 2025. Front Immunol. 2025. PMID: 40242761 Free PMC article.
References
-
- Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al.. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–45. Epub 2020/09/06. doi: 10.1038/s41590-020-0782-6 ; PubMed Central PMCID: PMC7611020. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

