LANCE: a Label-Free Live Apoptotic and Necrotic Cell Explorer Using Convolutional Neural Network Image Analysis
- PMID: 36251981
- PMCID: PMC10729583
- DOI: 10.1021/acs.analchem.2c00878
LANCE: a Label-Free Live Apoptotic and Necrotic Cell Explorer Using Convolutional Neural Network Image Analysis
Abstract
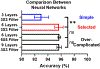
Identifying and quantifying cell death is the basis for all cell death research. Current methods for obtaining these quantitative measurements rely on established biomarkers, yet the marker-based approach suffers from limited marker specificity, high cost of reagents, lengthy sample preparation, and fluorescence imaging. Based on the morphological difference, we developed a Live, Apoptotic, and Necrotic Cell Explorer (LANCE) to categorize cell death status in a label-free manner, by incorporating machine learning and image processing. The LANCE workflow includes cropping individual cells from microscopic images having hundreds of cells, formation of an image database of around 5000 events, training and validation of the convolutional neural network models using multiple cell lines, and treatment conditions. With LANCE, we precisely categorized live, apoptotic, and necrotic cells with a high accuracy of 96.3 ± 0.5%. More importantly, the nondestructive label-free LANCE method allows for tracking time dynamics of the cell death process, which enhances the understanding of subtle cell death regulation at the molecular level. Hence, LANCE is a fast, low-cost, and nondestructive label-free method to distinguish cell status, which can be applied to cell death studies as well as many other biomedical applications.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures






References
-
- Martin SJ; Reutelingsperger CP; McGahon AJ; Rader JA; van Schie RC; LaFace DM; Green DR Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med 1995, 182, 1545–1556, DOI: 10.1084/jem.182.5.1545. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

