Site-to-site cross-talk in OST-B glycosylation of hCEACAM1-IgV
- PMID: 36251991
- PMCID: PMC9618145
- DOI: 10.1073/pnas.2202992119
Site-to-site cross-talk in OST-B glycosylation of hCEACAM1-IgV
Abstract
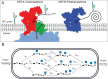
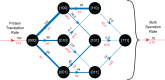
N-glycosylation is a common posttranslational modification of secreted proteins in eukaryotes. This modification targets asparagine residues within the consensus sequence, N-X-S/T. While this sequence is required for glycosylation, the initial transfer of a high-mannose glycan by oligosaccharyl transferases A or B (OST-A or OST-B) can lead to incomplete occupancy at a given site. Factors that determine the extent of transfer are not well understood, and understanding them may provide insight into the function of these important enzymes. Here, we use mass spectrometry (MS) to simultaneously measure relative occupancies for three N-glycosylation sites on the N-terminal IgV domain of the recombinant glycoprotein, hCEACAM1. We demonstrate that addition is primarily by the OST-B enzyme and propose a kinetic model of OST-B N-glycosylation. Fitting the kinetic model to the MS data yields distinct rates for glycan addition at most sites and suggests a largely stochastic initial order of glycan addition. The model also suggests that glycosylation at one site influences the efficiency of subsequent modifications at the other sites, and glycosylation at the central or N-terminal site leads to dead-end products that seldom lead to full glycosylation of all three sites. Only one path of progressive glycosylation, one initiated by glycosylation at the C-terminal site, can efficiently lead to full occupancy for all three sites. Thus, the hCEACAM1 domain provides an effective model system to study site-specific recognition of glycosylation sequons by OST-B and suggests that the order and efficiency of posttranslational glycosylation is influenced by steric cross-talk between adjoining acceptor sites.
Keywords: glycobiology; mass spectrometry; oligosaccharyltransferase.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Apweiler R., Hermjakob H., Sharon N., On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 (1999). - PubMed
-
- Hart G. W., Housley M. P., Slawson C., Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 (2007). - PubMed
-
- Stanley P., Taniguchi N., Aebi M., “N-Glycans” in Essentials of Glycobiology, Varki A., et al., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2017), chap. 9.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

