Nasopharyngeal microbiome of COVID-19 patients revealed a distinct bacterial profile in deceased and recovered individuals
- PMID: 36252893
- PMCID: PMC9568276
- DOI: 10.1016/j.micpath.2022.105829
Nasopharyngeal microbiome of COVID-19 patients revealed a distinct bacterial profile in deceased and recovered individuals
Abstract
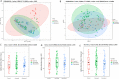
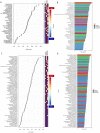
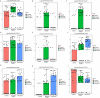
The bacterial co-infections in SARS-CoV-2 patients remained the least explored subject of clinical manifestations that may also determine the disease severity. Nasopharyngeal microbial community structure within SARS-CoV-2 infected patients could reveal interesting microbiome dynamics that may influence the disease outcomes. Here, in this research study, we analyzed distinct nasopharyngeal microbiome profile in the deceased (n = 48) and recovered (n = 29) COVID-19 patients and compared it with control SARS-CoV-2 negative individuals (control) (n = 33). The nasal microbiome composition of the three groups varies significantly (PERMANOVA, p-value <0.001), where deceased patients showed higher species richness compared to the recovered and control groups. Pathogenic genera, including Corynebacterium (LDA score 5.51), Staphylococcus, Serratia, Klebsiella and their corresponding species were determined as biomarkers (p-value <0.05, LDA cutoff 4.0) in the deceased COVID-19 patients. Ochrobactrum (LDA score 5.79), and Burkholderia (LDA 5.29), were found in the recovered group which harbors ordinal bacteria (p-value <0.05, LDA-4.0) as biomarkers. Similarly, Pseudomonas (LDA score 6.19), and several healthy nasal cavity commensals including Veillonella, and Porphyromonas, were biomarkers for the control individuals. Healthy commensal bacteria may trigger the immune response and alter the viral infection susceptibility and thus, may play important role and possible recovery that needs to be further explored. This research finding provide vital information and have significant implications for understanding the microbial diversity of COVID-19 patients. However, additional studies are needed to address the microbiome-based therapeutics and diagnostics interventions.
Keywords: And SARS-CoV-2; Nasopharyngeal microbiome; Opportunistic pathogens.
Copyright © 2022. Published by Elsevier Ltd.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Professor Chaitanya G Joshi reports financial support was provided by Government of Gujarat Department of Science and Technology.
Figures



Similar articles
-
SARS-CoV-2 infection reduces human nasopharyngeal commensal microbiome with inclusion of pathobionts.Sci Rep. 2021 Dec 15;11(1):24042. doi: 10.1038/s41598-021-03245-4. Sci Rep. 2021. PMID: 34911967 Free PMC article.
-
Metagenomic next-generation sequencing of nasopharyngeal microbiota in COVID-19 patients with different disease severities.Microbiol Spectr. 2024 May 2;12(5):e0416623. doi: 10.1128/spectrum.04166-23. Epub 2024 Apr 1. Microbiol Spectr. 2024. PMID: 38557102 Free PMC article.
-
Age-Related Changes in the Nasopharyngeal Microbiome Are Associated With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection and Symptoms Among Children, Adolescents, and Young Adults.Clin Infect Dis. 2022 Aug 24;75(1):e928-e937. doi: 10.1093/cid/ciac184. Clin Infect Dis. 2022. PMID: 35247047 Free PMC article.
-
Review article: Probiotics, prebiotics and dietary approaches during COVID-19 pandemic.Trends Food Sci Technol. 2021 Feb;108:187-196. doi: 10.1016/j.tifs.2020.12.009. Epub 2020 Dec 14. Trends Food Sci Technol. 2021. PMID: 33519087 Free PMC article. Review.
-
Autoimmunity and COVID-19 - The microbiotal connection.Autoimmun Rev. 2021 Aug;20(8):102865. doi: 10.1016/j.autrev.2021.102865. Epub 2021 Jun 10. Autoimmun Rev. 2021. PMID: 34118455 Free PMC article. Review.
Cited by
-
Oral Microbiota Alterations in Subjects with SARS-CoV-2 Displaying Prevalence of the Opportunistic Fungal Pathogen Candida albicans.Microorganisms. 2024 Jul 2;12(7):1356. doi: 10.3390/microorganisms12071356. Microorganisms. 2024. PMID: 39065125 Free PMC article.
-
Oropharyngeal Microbiome Analysis in Patients with Varying SARS-CoV-2 Infection Severity: A Prospective Cohort Study.J Pers Med. 2024 Mar 29;14(4):369. doi: 10.3390/jpm14040369. J Pers Med. 2024. PMID: 38672996 Free PMC article.
-
A cross-sectional study on the nasopharyngeal microbiota of individuals with SARS-CoV-2 infection across three COVID-19 waves in India.Front Microbiol. 2023 Sep 6;14:1238829. doi: 10.3389/fmicb.2023.1238829. eCollection 2023. Front Microbiol. 2023. PMID: 37744900 Free PMC article.
-
Analysis of the nasopharyngeal microbiome and respiratory pathogens in COVID-19 patients from Saudi Arabia.J Infect Public Health. 2023 May;16(5):680-688. doi: 10.1016/j.jiph.2023.03.001. Epub 2023 Mar 4. J Infect Public Health. 2023. PMID: 36934642 Free PMC article.
-
Microbiota Profile of the Nasal Cavity According to Lifestyles in Healthy Adults in Santiago, Chile.Microorganisms. 2023 Jun 22;11(7):1635. doi: 10.3390/microorganisms11071635. Microorganisms. 2023. PMID: 37512807 Free PMC article.
References
-
- Assessment R.R. 2021. Assessment of the Further Emergence and Potential Impact of the SARS-CoV-2 Omicron Variant of Concern in the Context of Ongoing Transmission of the Delta Variant of Concern in the EU/EEA, 18th Update.
-
- Chaudhari A.M., Joshi M., Kumar D., Patel A., Lokhande K.B., Krishnan A., Hanack K., Filipek S., Liepmann D., Renugopalakrishnan V., Paulmurugan R., Joshi C. Evaluation of immune evasion in SARS-CoV-2 Delta and Omicron variants. Comput. Struct. Biotechnol. J. 2022;20:4501–4516. doi: 10.1016/j.csbj.2022.08.010. - DOI - PMC - PubMed
-
- Liu J., Liu S., Zhang Z., Lee X., Wu W., Huang Z., Lei Z., Xu W., Chen D., Wu X., Guo Y., Peng L., Lin B., Chong Y., Mou X., Shi M., Lan P., Chen T., Zhao W., Gao Z. Association between the nasopharyngeal microbiome and metabolome in patients with COVID-19. Synth. Syst. Biotechnol. 2021;6:135–143. doi: 10.1016/j.synbio.2021.06.002. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

