A small molecule inhibitor prevents gut bacterial genotoxin production
- PMID: 36253549
- PMCID: PMC9889270
- DOI: 10.1038/s41589-022-01147-8
A small molecule inhibitor prevents gut bacterial genotoxin production
Abstract
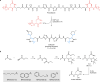
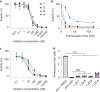
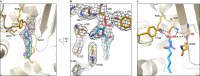
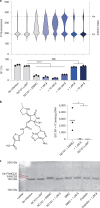
The human gut bacterial genotoxin colibactin is a possible key driver of colorectal cancer (CRC) development. Understanding colibactin's biological effects remains difficult owing to the instability of the proposed active species and the complexity of the gut microbiota. Here, we report small molecule boronic acid inhibitors of colibactin biosynthesis. Designed to mimic the biosynthetic precursor precolibactin, these compounds potently inhibit the colibactin-activating peptidase ClbP. Using biochemical assays and crystallography, we show that they engage the ClbP binding pocket, forming a covalent bond with the catalytic serine. These inhibitors reproduce the phenotypes observed in a clbP deletion mutant and block the genotoxic effects of colibactin on eukaryotic cells. The availability of ClbP inhibitors will allow precise, temporal control over colibactin production, enabling further study of its contributions to CRC. Finally, application of our inhibitors to related peptidase-encoding pathways highlights the power of chemical tools to probe natural product biosynthesis.
© 2022. The Author(s).
Conflict of interest statement
E.P.B. and M.R.V. are listed as inventors on a provisional patent (US application 63/135,825) that relates to the methods and compounds described here. The remaining authors declare no competing interests.
Figures

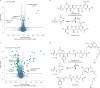
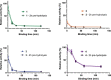
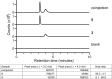


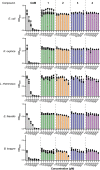
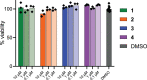
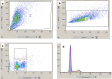
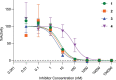

References
-
- Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019;16:690–704. - PubMed
-
- Janney A, Powrie F, Mann EH. Host-microbiome maladaptation in colorectal cancer. Nature. 2020;585:509–517. - PubMed
-
- Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

