Identification and differential usage of a host metalloproteinase entry pathway by SARS-CoV-2 Delta and Omicron
- PMID: 36254158
- PMCID: PMC9549715
- DOI: 10.1016/j.isci.2022.105316
Identification and differential usage of a host metalloproteinase entry pathway by SARS-CoV-2 Delta and Omicron
Abstract
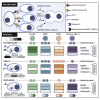
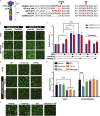
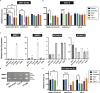
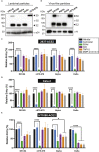
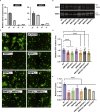
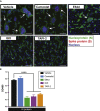
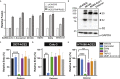
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike glycoprotein (S) binds to angiotensin-converting enzyme 2 (ACE2) to mediate membrane fusion via two distinct pathways: 1) a surface, serine protease-dependent or 2) an endosomal, cysteine protease-dependent pathway. In this study, we found that SARS-CoV-2 S has a wider protease usage and can also be activated by TMPRSS13 and matrix metalloproteinases (MMPs). We found that MMP-2 and MMP-9 played roles in SARS-CoV-2 S cell-cell fusion and TMPRSS2- and cathepsin-independent viral entry in cells expressing high MMP levels. MMP-dependent viral entry required cleavage at the S1/S2 junction in viral producer cells, and differential processing of variants of concern S dictated its usage; the efficiently processed Delta S preferred metalloproteinase-dependent entry when available, and less processed Omicron S was unable to us metalloproteinases for entry. As MMP-2/9 are released during inflammation, they may play roles in S-mediated cytopathic effects, tropism, and disease outcome.
Keywords: Biological sciences; microbiology; molecular biology; virology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity.J Virol. 2022 Apr 27;96(8):e0012822. doi: 10.1128/jvi.00128-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343766 Free PMC article.
-
Impact of SARS-CoV-2 Spike Mutations on Its Activation by TMPRSS2 and the Alternative TMPRSS13 Protease.mBio. 2022 Aug 30;13(4):e0137622. doi: 10.1128/mbio.01376-22. Epub 2022 Aug 1. mBio. 2022. PMID: 35913162 Free PMC article.
-
Metalloproteinase-Dependent and TMPRSS2-Independent Cell Surface Entry Pathway of SARS-CoV-2 Requires the Furin Cleavage Site and the S2 Domain of Spike Protein.mBio. 2022 Aug 30;13(4):e0051922. doi: 10.1128/mbio.00519-22. Epub 2022 Jun 16. mBio. 2022. PMID: 35708281 Free PMC article.
-
Proteolytic activation of SARS-CoV-2 spike protein.Microbiol Immunol. 2022 Jan;66(1):15-23. doi: 10.1111/1348-0421.12945. Epub 2021 Oct 12. Microbiol Immunol. 2022. PMID: 34561887 Free PMC article. Review.
-
Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: A systematic review of in vitro and in vivo studies.J Cell Physiol. 2021 Apr;236(4):2364-2392. doi: 10.1002/jcp.30032. Epub 2020 Sep 9. J Cell Physiol. 2021. PMID: 32901936
Cited by
-
SARS-CoV-2 and its impact on the cardiovascular and digestive systems - The interplay between new virus variants and human cells.Comput Struct Biotechnol J. 2023;21:1022-1029. doi: 10.1016/j.csbj.2023.01.024. Epub 2023 Jan 19. Comput Struct Biotechnol J. 2023. PMID: 36694807 Free PMC article. Review.
-
Development of ketobenzothiazole-based peptidomimetic TMPRSS13 inhibitors with low nanomolar potency.J Enzyme Inhib Med Chem. 2025 Dec;40(1):2466841. doi: 10.1080/14756366.2025.2466841. Epub 2025 Feb 20. J Enzyme Inhib Med Chem. 2025. PMID: 39976239 Free PMC article.
-
A Recent SARS-CoV-2 Infection Enhances Antibody-Dependent Cellular Cytotoxicity against Several Omicron Subvariants following a Fourth mRNA Vaccine Dose.Viruses. 2023 May 29;15(6):1274. doi: 10.3390/v15061274. Viruses. 2023. PMID: 37376574 Free PMC article.
-
Longitudinal humoral immunity against SARS-CoV-2 Spike following infection in individuals from Cameroon.Virology. 2025 Apr;605:110467. doi: 10.1016/j.virol.2025.110467. Epub 2025 Feb 25. Virology. 2025. PMID: 40037139
-
Genome-wide bioinformatics analysis of human protease capacity for proteolytic cleavage of the SARS-CoV-2 spike glycoprotein.Microbiol Spectr. 2024 Feb 6;12(2):e0353023. doi: 10.1128/spectrum.03530-23. Epub 2024 Jan 8. Microbiol Spectr. 2024. PMID: 38189333 Free PMC article.
References
-
- Abers M.S., Delmonte O.M., Ricotta E.E., Fintzi J., Fink D.L., De Jesus A.A.A., Zarember K.A., Alehashemi S., Oikonomou V., Desai J.V., et al. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6:e144455. doi: 10.1172/jci.insight.144455. - DOI - PMC - PubMed
-
- Arora P., Sidarovich A., Krüger N., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., Schulz S., Jäck H.M., et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021;37:109825. doi: 10.1016/j.celrep.2021.109825. - DOI - PMC - PubMed
-
- Avila-Mesquita C.D., Couto A.E.S., Campos L.C.B., Vasconcelos T.F., Michelon-Barbosa J., Corsi C.A.C., Mestriner F., Petroski-Moraes B.C., Garbellini-Diab M.J., Couto D.M.S., et al. MMP-2 and MMP-9 levels in plasma are altered and associated with mortality in COVID-19 patients. Biomed. Pharmacother. 2021;142:112067. doi: 10.1016/j.biopha.2021.112067. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

