A comprehensive mini-review on amyloidogenesis of different SARS-CoV-2 proteins and its effect on amyloid formation in various host proteins
- PMID: 36254263
- PMCID: PMC9558030
- DOI: 10.1007/s13205-022-03390-1
A comprehensive mini-review on amyloidogenesis of different SARS-CoV-2 proteins and its effect on amyloid formation in various host proteins
Abstract
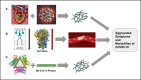
Amyloidogenesis is the inherent ability of proteins to change their conformation from native state to cross β-sheet rich fibrillar structures called amyloids which result in a wide range of diseases like Parkinson's disease, Alzheimer's disease, Finnish familial amyloidosis, ATTR amyloidosis, British and Danish dementia, etc. COVID-19, on the other hand is seen to have many similarities in symptoms with other amyloidogenic diseases and the overlap of these morbidities and symptoms led to the proposition whether SARS-CoV-2 proteins are undergoing amyloidogenesis and whether it is resulting in or aggravating amyloidogenesis of any human host protein. Thus the SARS-CoV-2 proteins in infected cells, i.e., Spike (S) protein, Nucleocapsid (N) protein, and Envelope (E) protein were tested via different machinery and amyloidogenesis in them were proven. In this review, we will analyze the pathway of amyloid formation in S-protein, N-protein, E-protein along with the effect that SARS-CoV-2 is creating on various host proteins leading to the unexpected onset of many morbidities like COVID-induced Acute Respiratory Distress Syndrome (ARDS), Parkinsonism in young COVID patients, formation of fibrin microthrombi in heart, etc., and their future implications.
Keywords: ARDS; ATTR; Amyloidogenesis; COVID-19; Envelope protein; Nucleocapsid protein; Serum amyloid A; Spike protein; α-Synuclein.
© King Abdulaziz City for Science and Technology 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interest.
Figures



Similar articles
-
Controversial Properties of Amyloidogenic Proteins and Peptides: New Data in the COVID Era.Biomedicines. 2023 Apr 19;11(4):1215. doi: 10.3390/biomedicines11041215. Biomedicines. 2023. PMID: 37189833 Free PMC article. Review.
-
SARS-COV-2 spike protein fragment eases amyloidogenesis of α-synuclein.J Chem Phys. 2023 Jul 7;159(1):015103. doi: 10.1063/5.0157331. J Chem Phys. 2023. PMID: 37409768 Free PMC article.
-
Amyloidogenesis of SARS-CoV-2 Spike Protein.J Am Chem Soc. 2022 May 25;144(20):8945-8950. doi: 10.1021/jacs.2c03925. Epub 2022 May 17. J Am Chem Soc. 2022. PMID: 35579205 Free PMC article.
-
Interactions between SARS-CoV-2 N-Protein and α-Synuclein Accelerate Amyloid Formation.ACS Chem Neurosci. 2022 Jan 5;13(1):143-150. doi: 10.1021/acschemneuro.1c00666. Epub 2021 Dec 3. ACS Chem Neurosci. 2022. PMID: 34860005 Free PMC article.
-
Exploring critical determinants of protein amyloidogenesis: a review.J Pept Sci. 2013 Sep;19(9):529-36. doi: 10.1002/psc.2539. Epub 2013 Jul 19. J Pept Sci. 2013. PMID: 23873708 Review.
Cited by
-
Host-Pathogen Interactions Influencing Zoonotic Spillover Potential and Transmission in Humans.Viruses. 2023 Feb 22;15(3):599. doi: 10.3390/v15030599. Viruses. 2023. PMID: 36992308 Free PMC article. Review.
-
Controversial Properties of Amyloidogenic Proteins and Peptides: New Data in the COVID Era.Biomedicines. 2023 Apr 19;11(4):1215. doi: 10.3390/biomedicines11041215. Biomedicines. 2023. PMID: 37189833 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

