Impact of Clinical Data Veracity on Cancer Genomic Research
- PMID: 36255250
- PMCID: PMC9648686
- DOI: 10.1093/jncics/pkac070
Impact of Clinical Data Veracity on Cancer Genomic Research
Abstract
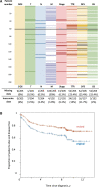
Genomic analysis of tumors is transforming our understanding of cancer. However, although a great deal of attention is paid to the accuracy of the cancer genomic data itself, less attention has been paid to the accuracy of the associated clinical information that renders the genomic data useful for research. In this brief communication, we suggest that omissions and errors in clinical annotations have a major impact on the interpretation of cancer genomic data. We describe our discovery of annotation omissions and errors when reviewing an already carefully annotated colorectal cancer gene expression dataset from our laboratory. The potential importance of clinical annotation omissions and errors was then explored using simulation analyses with an independent genomic dataset. We suggest that the completeness and veracity of clinical annotations accompanying cancer genomic data require renewed focus by the oncology research community, when planning new collections and when interpreting existing cancer genomic data.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
