Statistical Evaluation of Metaproteomics and 16S rRNA Amplicon Sequencing Techniques for Study of Gut Microbiota Establishment in Infants with Cystic Fibrosis
- PMID: 36255300
- PMCID: PMC9784762
- DOI: 10.1128/spectrum.01466-22
Statistical Evaluation of Metaproteomics and 16S rRNA Amplicon Sequencing Techniques for Study of Gut Microbiota Establishment in Infants with Cystic Fibrosis
Abstract
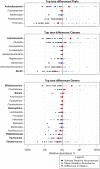
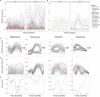
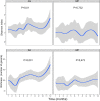
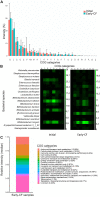
Newborn screening for cystic fibrosis (CF) can identify affected but asymptomatic infants. The selection of omic technique for gut microbiota study is crucial due to both the small amount of feces available and the low microorganism load. Our aims were to compare the agreement between 16S rRNA amplicon sequencing and metaproteomics by a robust statistical analysis, including both presence and abundance of taxa, to describe the sequential establishment of the gut microbiota during the first year of life in a small size sample (8 infants and 28 fecal samples). The taxonomic assignations by the two techniques were similar, whereas certain discrepancies were observed in the abundance detection, mostly the lower predicted relative abundance of Bifidobacterium and the higher predicted relative abundance of certain Firmicutes and Proteobacteria by amplicon sequencing. During the first months of life, the CF gut microbiota is characterized by a significant enrichment of Ruminococcus gnavus, the expression of certain virulent bacterial traits, and the detection of human inflammation-related proteins. Metaproteomics provides information on composition and functionality, as well as data on host-microbiome interactions. Its strength is the identification and quantification of Actinobacteria and certain classes of Firmicutes, but alpha diversity indices are not comparable to those of amplicon sequencing. Both techniques detected an aberrant microbiota in our small cohort of infants with CF during their first year of life, dominated by the enrichment of R. gnavus within a human inflammatory environment. IMPORTANCE In recent years, some techniques have been incorporated for the study of microbial ecosystems, being 16S rRNA gene sequencing being the most widely used. Metaproteomics provides the advantage of identifying the interaction between microorganisms and human cells, but the available databases are less extensive as well as imprecise. Few studies compare the statistical differences between the two techniques to define the composition of an ecosystem. Our work shows that the two methods are comparable in terms of microorganism identification but provide different results in alpha diversity analysis. On the other hand, we have studied newborns with cystic fibrosis, for whom we have described the establishment of an intestinal ecosystem marked by the inflammatory response of the host and the enrichment of Ruminococcus gnavus.
Keywords: Bland-Altman test; Ruminococcus gnavus; amplicon sequencing; cystic fibrosis; gut microbiota establishment; metaproteomics.
Conflict of interest statement
The authors declare a conflict of interest. Rosa del Campo is recipient of a Vertex grant. The remaining authors have no conflict of interest.
Figures


References
-
- Selma-Royo M, Calatayud Arroyo M, García-Mantrana I, Parra-Llorca A, Escuriet R, Martínez-Costa C, Collado MC. 2020. Perinatal environment shapes microbiota colonization and infant growth: impact on host response and intestinal function. Microbiome 8:167. doi: 10.1186/s40168-020-00940-8. - DOI - PMC - PubMed
-
- Cuthbertson L, Walker AW, Oliver AE, Rogers GB, Rivett DW, Hampton TH, Ashare A, Elborn JS, De Soyza A, Carroll MP, Hoffman LR, Lanyon C, Moskowitz SM, O’Toole GA, Parkhill J, Planet PJ, Teneback CC, Tunney MM, Zuckerman JB, Bruce KD, van der Gast CJ. 2020. Lung function and microbiota diversity in cystic fibrosis. Microbiome 8:45. doi: 10.1186/s40168-020-00810-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

