Discovery of a MUC3B gene reconstructs the membrane mucin gene cluster on human chromosome 7
- PMID: 36256656
- PMCID: PMC9578598
- DOI: 10.1371/journal.pone.0275671
Discovery of a MUC3B gene reconstructs the membrane mucin gene cluster on human chromosome 7
Abstract
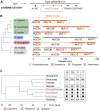
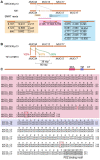
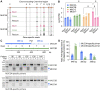
Human tissue surfaces are coated with mucins, a family of macromolecular sugar-laden proteins serving diverse functions from lubrication to the formation of selective biochemical barriers against harmful microorganisms and molecules. Membrane mucins are a distinct group of mucins that are attached to epithelial cell surfaces where they create a dense glycocalyx facing the extracellular environment. All mucin proteins carry long stretches of tandemly repeated sequences that undergo extensive O-linked glycosylation to form linear mucin domains. However, the repetitive nature of mucin domains makes them prone to recombination and renders their genetic sequences particularly difficult to read with standard sequencing technologies. As a result, human mucin genes suffer from significant sequence gaps that have hampered the investigation of gene function in health and disease. Here we leveraged a recent human genome assembly to characterize a previously unmapped MUC3B gene located at the q22 locus on chromosome 7, within a cluster of four structurally related membrane mucin genes that we name the MUC3 cluster. We found that MUC3B shares high sequence identity with the known MUC3A gene and that the two genes are governed by evolutionarily conserved regulatory elements. Furthermore, we show that MUC3A, MUC3B, MUC12, and MUC17 in the human MUC3 cluster are expressed in intestinal epithelial cells (IECs). Our results complete existing genetic gaps in the MUC3 cluster which is a conserved genetic unit in vertebrates. We anticipate our results to be the starting point for the detection of disease-associated polymorphisms in the human MUC3 cluster. Moreover, our study provides the basis for the exploration of intestinal mucin gene function in widely used experimental models such as human intestinal organoids and genetic mouse models.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Initiation of transcription of the MUC3A human intestinal mucin from a TATA-less promoter and comparison with the MUC3B amino terminus.J Biol Chem. 2003 Dec 5;278(49):49600-9. doi: 10.1074/jbc.M305769200. Epub 2003 Sep 4. J Biol Chem. 2003. PMID: 12958310
-
MUC17, a novel membrane-tethered mucin.Biochem Biophys Res Commun. 2002 Mar 1;291(3):466-75. doi: 10.1006/bbrc.2002.6475. Biochem Biophys Res Commun. 2002. PMID: 11855812
-
Activity of recombinant cysteine-rich domain proteins derived from the membrane-bound MUC17/Muc3 family mucins.Biochim Biophys Acta. 2010 Jul;1800(7):629-38. doi: 10.1016/j.bbagen.2010.03.010. Epub 2010 Mar 20. Biochim Biophys Acta. 2010. PMID: 20332014 Free PMC article.
-
Intestinal goblet cells and mucins in health and disease: recent insights and progress.Curr Gastroenterol Rep. 2010 Oct;12(5):319-30. doi: 10.1007/s11894-010-0131-2. Curr Gastroenterol Rep. 2010. PMID: 20703838 Free PMC article. Review.
-
Architecture of the large membrane-bound mucins.Gene. 2008 Mar 15;410(2):215-22. doi: 10.1016/j.gene.2007.12.014. Epub 2007 Dec 28. Gene. 2008. PMID: 18242885 Review.
Cited by
-
MUC17 is an essential small intestinal glycocalyx component that is disrupted in Crohn's disease.bioRxiv [Preprint]. 2024 Feb 12:2024.02.08.578867. doi: 10.1101/2024.02.08.578867. bioRxiv. 2024. Update in: JCI Insight. 2024 Dec 19;10(3):e181481. doi: 10.1172/jci.insight.181481. PMID: 38405862 Free PMC article. Updated. Preprint.
-
MUC17 is an essential small intestinal glycocalyx component that is disrupted in Crohn's disease.JCI Insight. 2024 Dec 19;10(3):e181481. doi: 10.1172/jci.insight.181481. JCI Insight. 2024. PMID: 39699961 Free PMC article.
-
Transcriptome driven discovery of novel candidate genes for human neurological disorders in the telomer-to-telomer genome assembly era.Hum Genomics. 2023 Oct 23;17(1):94. doi: 10.1186/s40246-023-00543-y. Hum Genomics. 2023. PMID: 37872607 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

