Reprogramming of myeloid cells and their progenitors in patients with non-medullary thyroid carcinoma
- PMID: 36257966
- PMCID: PMC9579179
- DOI: 10.1038/s41467-022-33907-4
Reprogramming of myeloid cells and their progenitors in patients with non-medullary thyroid carcinoma
Abstract
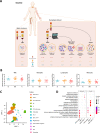
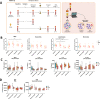
Myeloid cells, crucial players in antitumoral defense, are affected by tumor-derived factors and treatment. The role of myeloid cells and their progenitors prior to tumor infiltration is poorly understood. Here we show single-cell transcriptomics and functional analyses of the myeloid cell lineage in patients with non-medullary thyroid carcinoma (TC) and multinodular goiter, before and after treatment with radioactive iodine compared to healthy controls. Integrative data analysis indicates that monocytes of TC patients have transcriptional upregulation of antigen presentation, reduced cytokine production capacity, and overproduction of reactive oxygen species. Interestingly, these cancer-related pathological changes are partially removed upon treatment. In bone marrow, TC patients tend to shift from myelopoiesis towards lymphopoiesis, reflected in transcriptional differences. Taken together, distinct transcriptional and functional changes in myeloid cells arise before their infiltration of the tumor and are already initiated in bone marrow, which suggests an active role in forming the tumor immune microenvironment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

