Prognostic risk assessment model and drug sensitivity analysis of colon adenocarcinoma (COAD) based on immune-related lncRNA pairs
- PMID: 36258178
- PMCID: PMC9579580
- DOI: 10.1186/s12859-022-04969-4
Prognostic risk assessment model and drug sensitivity analysis of colon adenocarcinoma (COAD) based on immune-related lncRNA pairs
Abstract
Purpose: The aim of this study was to identify and screen long non-coding RNA (lncRNA) associated with immune genes in colon cancer, construct immune-related lncRNA pairs, establish a prognostic risk assessment model for colon adenocarcinoma (COAD), and explore prognostic factors and drug sensitivity.
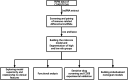
Method: Our method was based on data from The Cancer Genome Atlas (TCGA). To begin, we obtained all pertinent demographic and clinical information on 385 patients with COAD. All lncRNAs significantly related to immune genes and with differential expression were identified to construct immune lncRNA pairs. Subsequently, least absolute shrinkage and selection operator and Cox models were used to screen out prognostic-related immune lncRNAs for the establishment of a prognostic risk scoring formula. Finally, We analysed the functional differences between subgroups and screened the drugs, and establish an individual prediction nomogram model.
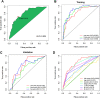
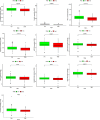
Results: Our final analysis confirmed eight lncRNA pairs to construct prognostic risk assessment model. Results showed that the high-risk and low-risk groups had significant differences (training (n = 249): p < 0.001, validation (n = 114): p = 0.022). The prognostic model was certified as an independent prognosis model. Compared with the common clinicopathological indicators, the prognostic model had better predictive efficiency (area under the curve (AUC) = 0.805). Finally, We have analysed highly differentiated cellular pathways such as mucosal immune response, identified 9 differential immune cells, 10 sensitive drugs, and establish an individual prediction nomogram model (C-index = 0.820).
Conclusion: Our study verified that the eight lncRNA pairs mentioned can be used as biomarkers to predict the prognosis of COAD patients. Identified cells, drugs may have an positive effect on colon cancer prognosis.
Keywords: Colon adenocarcinoma; Drug sensitivity analysis; Immune-related lncRNA pairs; Prognostic risk assessment model.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

