OPAQUE3, encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice
- PMID: 36258666
- PMCID: PMC9700205
- DOI: 10.1016/j.xplc.2022.100463
OPAQUE3, encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice
Abstract
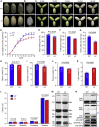
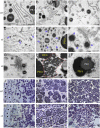
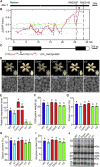
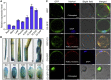
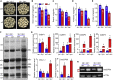
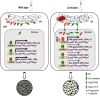
Starch and storage proteins are the main components of rice (Oryza sativa L.) grains. Despite their importance, the molecular regulatory mechanisms of storage protein and starch biosynthesis remain largely elusive. Here, we identified a rice opaque endosperm mutant, opaque3 (o3), that overaccumulates 57-kDa proglutelins and has significantly lower protein and starch contents than the wild type. The o3 mutant also has abnormal protein body structures and compound starch grains in its endosperm cells. OPAQUE3 (O3) encodes a transmembrane basic leucine zipper (bZIP) transcription factor (OsbZIP60) and is localized in the endoplasmic reticulum (ER) and the nucleus, but it is localized mostly in the nucleus under ER stress. We demonstrated that O3 could activate the expression of several starch synthesis-related genes (GBSSI, AGPL2, SBEI, and ISA2) and storage protein synthesis-related genes (OsGluA2, Prol14, and Glb1). O3 also plays an important role in protein processing and export in the ER by directly binding to the promoters and activating the expression of OsBIP1 and PDIL1-1, two major chaperones that assist with folding of immature secretory proteins in the ER of rice endosperm cells. High-temperature conditions aggravate ER stress and result in more abnormal grain development in o3 mutants. We also revealed that OsbZIP50 can assist O3 in response to ER stress, especially under high-temperature conditions. We thus demonstrate that O3 plays a central role in rice grain development by participating simultaneously in the regulation of storage protein and starch biosynthesis and the maintenance of ER homeostasis in endosperm cells.
Keywords: ER stress; OPAQUE3; grain development; high temperature; rice.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., Yoshida K., Mitsuoka C., Tamiru M., et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012;30:174–178. - PubMed
-
- Ball S.G., Morell M.K. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003;54:207–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

