Ribovirus classification by a polymerase barcode sequence
- PMID: 36258794
- PMCID: PMC9573346
- DOI: 10.7717/peerj.14055
Ribovirus classification by a polymerase barcode sequence
Erratum in
-
Correction: Ribovirus classification by a polymerase barcode sequence.PeerJ. 2024 Aug 9;10:e14055/correction-1. doi: 10.7717/peerj.14055/correction-1. eCollection 2024. PeerJ. 2024. PMID: 39139825 Free PMC article.
Abstract
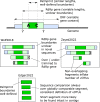
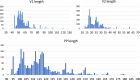
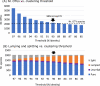
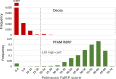
RNA viruses encoding a polymerase gene (riboviruses) dominate the known eukaryotic virome. High-throughput sequencing is revealing a wealth of new riboviruses known only from sequence, precluding classification by traditional taxonomic methods. Sequence classification is often based on polymerase sequences, but standardised methods to support this approach are currently lacking. To address this need, we describe the polymerase palmprint, a segment of the palm sub-domain robustly delineated by well-conserved catalytic motifs. We present an algorithm, Palmscan, which identifies palmprints in nucleotide and amino acid sequences; PALMdb, a collection of palmprints derived from public sequence databases; and palmID, a public website implementing palmprint identification, search, and annotation. Together, these methods demonstrate a proof-of-concept workflow for high-throughput characterisation of RNA viruses, paving the path for the continued rapid growth in RNA virus discovery anticipated in the coming decade.
Keywords: RNA virus; RNA-dependent RNA polymerase; Virus classification; Virus evolution.
© 2022 Babaian and Edgar.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Abarenkov K, Henrik Nilsson R, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytologist. 2010;186(2):281–285. doi: 10.1111/nph.2010.186.issue-2. - DOI - PubMed
-
- Bennett AJ, Paskey AC, Ebinger A, Pfaff F, Priemer G, Höper D, Breithaupt A, Heuser E, Ulrich RG, Kuhn JH, Bishop-Lilly KA, Beer M, Goldberg TL. Relatives of rubella virus in diverse mammals. Nature. 2020;586:424–428. doi: 10.1038/s41586-020-2812-9. Number: 7829 Publisher: Nature Publishing Group. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

