Host genetic requirements for DNA release of lactococcal phage TP901-1
- PMID: 36259418
- PMCID: PMC9733650
- DOI: 10.1111/1751-7915.14156
Host genetic requirements for DNA release of lactococcal phage TP901-1
Abstract
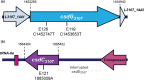
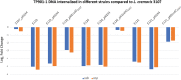
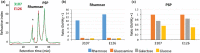
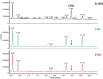
The first step in phage infection is the recognition of, and adsorption to, a receptor located on the host cell surface. This reversible host adsorption step is commonly followed by an irreversible event, which involves phage DNA delivery or release into the bacterial cytoplasm. The molecular components that trigger this latter event are unknown for most phages of Gram-positive bacteria. In the current study, we present a comparative genome analysis of three mutants of Lactococcus cremoris 3107, which are resistant to the P335 group phage TP901-1 due to mutations that affect TP901-1 DNA release. Through genetic complementation and phage infection assays, a predicted lactococcal three-component glycosylation system (TGS) was shown to be required for TP901-1 infection. Major cell wall saccharidic components were analysed, but no differences were found. However, heterologous gene expression experiments indicate that this TGS is involved in the glucosylation of a cell envelope-associated component that triggers TP901-1 DNA release. To date, a saccharide modification has not been implicated in the DNA delivery process of a Gram-positive infecting phage.
© 2022 The Authors. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. - PubMed
-
- Andres, D. , Roske, Y. , Doering, C. , Heinemann, U. , Seckler, R. & Barbirz, S. (2012) Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Molecular Microbiology, 83, 1244–1253. - PubMed
-
- Blatny, J.M. , Godager, L. , Lunde, M. & Nes, I.F. (2004) Complete genome sequence of the Lactococcus lactis temperate phage φLC3: comparative analysis of φLC3 and its relatives in lactococci and streptococci. Virology, 318, 231–244. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

