Comparative Genomic Insights into the Evolution of Halobacteria-Associated " Candidatus Nanohaloarchaeota"
- PMID: 36259734
- PMCID: PMC9765267
- DOI: 10.1128/msystems.00669-22
Comparative Genomic Insights into the Evolution of Halobacteria-Associated " Candidatus Nanohaloarchaeota"
Abstract
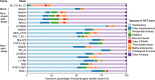
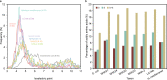
Members of the phylum "Candidatus Nanohaloarchaeota," a representative lineage within the DPANN superphylum, are characterized by their nanosized cells and symbiotic lifestyle with Halobacteria. However, the development of the symbiosis remains unclear. Here, we propose two novel families, "Candidatus Nanoanaerosalinaceae" and "Candidatus Nanohalalkaliarchaeaceae" in "Ca. Nanohaloarchaeota," represented by five dereplicated metagenome-assembled genomes obtained from hypersaline sediments or related enrichment cultures of soda-saline lakes. Phylogenetic analyses reveal that the two novel families are placed at the root of the family "Candidatus Nanosalinaceae," including the cultivated taxa. The two novel families prefer hypersaline sediments, and the acid shift of predicted proteomes indicates a "salt-in" strategy for hypersaline adaptation. They contain a lower proportion of putative horizontal gene transfers from Halobacteria than "Ca. Nanosalinaceae," suggesting a weaker association with Halobacteria. Functional prediction and historical events reconstruction disclose that they exhibit divergent potentials in carbohydrate and organic acid metabolism and environmental responses. Globally, comparative genomic analyses based on the new families enrich the taxonomic and functional diversity of "Ca. Nanohaloarchaeota" and provide insights into the evolutionary process of "Ca. Nanohaloarchaeota" and their symbiotic relationship with Halobacteria. IMPORTANCE The DPANN superphylum is a group of archaea widely distributed in various habitats. They generally have small cells and have a symbiotic lifestyle with other archaea. The archaeal symbiotic interaction is vital to understanding microbial communities. However, the formation and evolution of the symbiosis between the DPANN lineages and other diverse archaea remain unclear. Based on phylogeny, habitat distribution, hypersaline adaptation, host prediction, functional potentials, and historical events of "Ca. Nanohaloarchaeota," a representative phylum within the DPANN superphylum, we report two novel families representing intermediate stages, and we infer the evolutionary process of "Ca. Nanohaloarchaeota" and their Halobacteria-associated symbiosis. Altogether, this research helps in understanding the evolution of symbiosis in "Ca. Nanohaloarchaeota" and provides a model for the evolution of other DPANN lineages.
Keywords: DPANN superphylum; comparative genomics; evolution; horizontal gene transfer; symbiosis; “Candidatus Nanohaloarchaeota”.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi:10.1038/nature12352. - DOI - PubMed
-
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE. 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6:81–93. doi:10.1038/ismej.2011.78. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

