Genomic landscape of drug response reveals mediators of anthelmintic resistance
- PMID: 36261007
- PMCID: PMC9597552
- DOI: 10.1016/j.celrep.2022.111522
Genomic landscape of drug response reveals mediators of anthelmintic resistance
Abstract
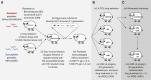
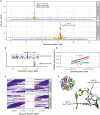
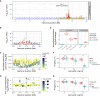
Like other pathogens, parasitic helminths can rapidly evolve resistance to drug treatment. Understanding the genetic basis of anthelmintic drug resistance in parasitic nematodes is key to tracking its spread and improving the efficacy and sustainability of parasite control. Here, we use an in vivo genetic cross between drug-susceptible and multi-drug-resistant strains of Haemonchus contortus in a natural host-parasite system to simultaneously map resistance loci for the three major classes of anthelmintics. This approach identifies new alleles for resistance to benzimidazoles and levamisole and implicates the transcription factor cky-1 in ivermectin resistance. This gene is within a locus under selection in ivermectin-resistant populations worldwide; expression analyses and functional validation using knockdown experiments support that cky-1 is associated with ivermectin survival. Our work demonstrates the feasibility of high-resolution forward genetics in a parasitic nematode and identifies variants for the development of molecular diagnostics to combat drug resistance in the field.
Keywords: CP: Microbiology; CP: Molecular biology; Haemonchus contortus; anthelmintic resistance; benzimidazole; cky-1; forward genetics; genetic cross; genome-wide association; helminth; ivermectin; levamisole.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Avramenko R.W., Redman E.M., Melville L., Bartley Y., Wit J., Queiroz C., Bartley D.J., Gilleard J.S. Deep amplicon sequencing as a powerful new tool to screen for sequence polymorphisms associated with anthelmintic resistance in parasitic nematode populations. Int. J. Parasitol. 2019;49:13–26. - PubMed
-
- Baker R.H., Buschbaum S., Matthews J.B., McKendrick I.J., Schnieder T., Strube C., Nisbet A.J. GTP-Cyclohydrolase and development in Teladorsagia circumcincta and Dictyocaulus viviparus (Nematoda: Strongylida) Exp. Parasitol. 2011;128:309–317. - PubMed
-
- Barrère V., Beech R.N., Charvet C.L., Prichard R.K. Novel assay for the detection and monitoring of levamisole resistance in Haemonchus contortus. Int. J. Parasitol. 2014;44:235–241. - PubMed
-
- Bartley D.J., Jackson F., Jackson E., Sargison N. Characterisation of two triple resistant field isolates of Teladorsagia from Scottish lowland sheep farms. Vet. Parasitol. 2004;123:189–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

