Activity-Induced Cortical Glutamatergic Neuron Nascent Proteins
- PMID: 36261270
- PMCID: PMC9617616
- DOI: 10.1523/JNEUROSCI.0707-22.2022
Activity-Induced Cortical Glutamatergic Neuron Nascent Proteins
Erratum in
-
Erratum: Schiapparelli et al., "Activity-Induced Cortical Glutamatergic Neuron Nascent Proteins".J Neurosci. 2023 Jun 21;43(25):4750. doi: 10.1523/JNEUROSCI.0903-23.2023. Epub 2023 May 30. J Neurosci. 2023. PMID: 37253604 Free PMC article. No abstract available.
Abstract
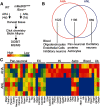
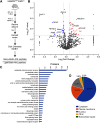
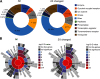
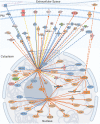
Neuronal activity initiates signaling cascades that culminate in diverse outcomes including structural and functional neuronal plasticity, and metabolic changes. While studies have revealed activity-dependent neuronal cell type-specific transcriptional changes, unbiased quantitative analysis of cell-specific activity-induced dynamics in newly synthesized proteins (NSPs) synthesis in vivo has been complicated by cellular heterogeneity and a relatively low abundance of NSPs within the proteome in the brain. Here we combined targeted expression of mutant MetRS (methionine tRNA synthetase) in genetically defined cortical glutamatergic neurons with tight temporal control of treatment with the noncanonical amino acid, azidonorleucine, to biotinylate NSPs within a short period after pharmacologically induced seizure in male and female mice. By purifying peptides tagged with heavy or light biotin-alkynes and using direct tandem mass spectrometry detection of biotinylated peptides, we quantified activity-induced changes in cortical glutamatergic neuron NSPs. Seizure triggered significant changes in ∼300 NSPs, 33% of which were decreased by seizure. Proteins mediating excitatory and inhibitory synaptic plasticity, including SynGAP1, Pak3, GEPH1, Copine-6, and collybistin, and DNA and chromatin remodeling proteins, including Rad21, Smarca2, and Ddb1, are differentially synthesized in response to activity. Proteins likely to play homeostatic roles in response to activity, such as regulators of proteastasis, intracellular ion control, and cytoskeleton remodeling proteins, are activity induced. Conversely, seizure decreased newly synthetized NCAM, among others, suggesting that seizure induced degradation. Overall, we identified quantitative changes in the activity-induced nascent proteome from genetically defined cortical glutamatergic neurons as a strategy to discover downstream mediators of neuronal plasticity and generate hypotheses regarding their function.SIGNIFICANCE STATEMENT Activity-induced neuronal and synaptic plasticity are mediated by changes in the protein landscape, including changes in the activity-induced newly synthesized proteins; however, identifying neuronal cell type-specific nascent proteome dynamics in the intact brain has been technically challenging. We conducted an unbiased proteomic screen from which we identified significant activity-induced changes in ∼300 newly synthesized proteins in genetically defined cortical glutamatergic neurons within 20 h after pharmacologically induced seizure. Bioinformatic analysis of the dynamic nascent proteome indicates that the newly synthesized proteins play diverse roles in excitatory and inhibitory synaptic plasticity, chromatin remodeling, homeostatic mechanisms, and proteasomal and metabolic functions, extending our understanding of the diversity of plasticity mechanisms.
Keywords: BONCAT; activity dependent; cortex; nascent protein; neuroproteomics; seizure.
Copyright © 2022 Schiapparelli et al.
Figures





References
-
- Alvarez-Castelao B, Schanzenbächer CT, Hanus C, Glock C, Tom Dieck S, Dörrbaum AR, Bartnik I, Nassim-Assir B, Ciirdaeva E, Mueller A, Dieterich DC, Tirrell DA, Langer JD, Schuman EM (2017) Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat Biotechnol 35:1196–1201. 10.1038/nbt.4016 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
