Borgs are giant genetic elements with potential to expand metabolic capacity
- PMID: 36261517
- PMCID: PMC9605863
- DOI: 10.1038/s41586-022-05256-1
Borgs are giant genetic elements with potential to expand metabolic capacity
Abstract
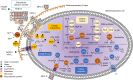
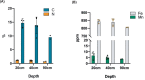
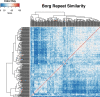
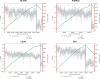
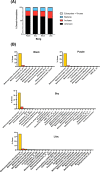
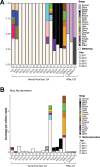
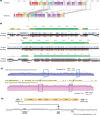
Anaerobic methane oxidation exerts a key control on greenhouse gas emissions1, yet factors that modulate the activity of microorganisms performing this function remain poorly understood. Here we discovered extraordinarily large, diverse DNA sequences that primarily encode hypothetical proteins through studying groundwater, sediments and wetland soil where methane production and oxidation occur. Four curated, complete genomes are linear, up to approximately 1 Mb in length and share genome organization, including replichore structure, long inverted terminal repeats and genome-wide unique perfect tandem direct repeats that are intergenic or generate amino acid repeats. We infer that these are highly divergent archaeal extrachromosomal elements with a distinct evolutionary origin. Gene sequence similarity, phylogeny and local divergence of sequence composition indicate that many of their genes were assimilated from methane-oxidizing Methanoperedens archaea. We refer to these elements as 'Borgs'. We identified at least 19 different Borg types coexisting with Methanoperedens spp. in four distinct ecosystems. Borgs provide methane-oxidizing Methanoperedens archaea access to genes encoding proteins involved in redox reactions and energy conservation (for example, clusters of multihaem cytochromes and methyl coenzyme M reductase). These data suggest that Borgs might have previously unrecognized roles in the metabolism of this group of archaea, which are known to modulate greenhouse gas emissions, but further studies are now needed to establish their functional relevance.
© 2022. The Author(s).
Conflict of interest statement
J.F.B. is a co-founder of Metagenomi. J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, Intellia Therapeutics and Mammoth Biosciences; is a scientific advisory board member of Vertex, Caribou Biosciences, Intellia Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Algen Biotechnologies, Felix Biosciences, The Column Group and Inari Agriculture; is Chief Science Advisor to Sixth Street, a director at Johnson & Johnson, Altos and Tempus; and has research projects sponsored by Apple Tree Partners and Roche.
Figures





Comment in
-
Mystery find of microbial DNA elements called Borgs.Nature. 2022 Oct;610(7933):635-637. doi: 10.1038/d41586-022-02975-3. Nature. 2022. PMID: 36261713 No abstract available.
References
-
- Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008;6:579–591. - PubMed
-
- Boetius A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

