Genetic insights into the social organization of Neanderthals
- PMID: 36261548
- PMCID: PMC9581778
- DOI: 10.1038/s41586-022-05283-y
Genetic insights into the social organization of Neanderthals
Abstract
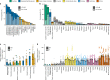
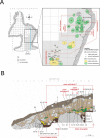
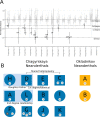
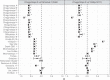
Genomic analyses of Neanderthals have previously provided insights into their population history and relationship to modern humans1-8, but the social organization of Neanderthal communities remains poorly understood. Here we present genetic data for 13 Neanderthals from two Middle Palaeolithic sites in the Altai Mountains of southern Siberia: 11 from Chagyrskaya Cave9,10 and 2 from Okladnikov Cave11-making this one of the largest genetic studies of a Neanderthal population to date. We used hybridization capture to obtain genome-wide nuclear data, as well as mitochondrial and Y-chromosome sequences. Some Chagyrskaya individuals were closely related, including a father-daughter pair and a pair of second-degree relatives, indicating that at least some of the individuals lived at the same time. Up to one-third of these individuals' genomes had long segments of homozygosity, suggesting that the Chagyrskaya Neanderthals were part of a small community. In addition, the Y-chromosome diversity is an order of magnitude lower than the mitochondrial diversity, a pattern that we found is best explained by female migration between communities. Thus, the genetic data presented here provide a detailed documentation of the social organization of an isolated Neanderthal community at the easternmost extent of their known range.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
The first genomic portrait of a Neanderthal family.Nature. 2022 Oct;610(7932):454-455. doi: 10.1038/d41586-022-03005-y. Nature. 2022. PMID: 36261544 No abstract available.
-
First known Neanderthal family discovered in Siberian cave.Nature. 2022 Oct;610(7933):615-616. doi: 10.1038/d41586-022-03339-7. Nature. 2022. PMID: 36261727 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

