SF3B1 mutated MDS: Blast count, genetic co-abnormalities and their impact on classification and prognosis
- PMID: 36261576
- PMCID: PMC9712089
- DOI: 10.1038/s41375-022-01728-5
SF3B1 mutated MDS: Blast count, genetic co-abnormalities and their impact on classification and prognosis
Abstract
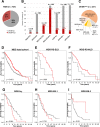
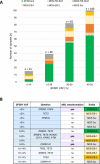
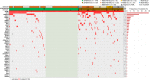
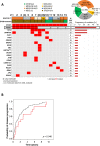
Recently, MDS with mutated SF3B1 and blast count <5% was proposed as distinct entity with favorable prognosis by the international working group for the prognosis of MDS (IWG-PM), the 5th edition of the WHO classification and the International Consensus Classification. To further characterize this entity with respect to the genomic landscape, AML transformation rate and clinical outcome, we analyzed 734 MDS patients by whole genome sequencing. SF3B1 mutations were identified in 31% (n = 231), most frequently accompanied by TET2 mutations (29%). 144/231 (62%) SF3B1mut samples fulfilled entity criteria proposed by IWG-PM (SF3B1ent). These cases were associated with longer survival, lower AML transformation rate, normal karyotypes and harbored less accompanying mutations compared to SF3B1mut samples not falling into the proposed SF3B1 entity (SF3B1nent). Of SF3B1mut cases 7% (15/231; SF3B1ent: 3/144 [2%]; SF3B1nent: 12/87 [14%]) progressed to AML compared to 15% SF3B1 wild-type patients (75/503). Of these 15 SF3B1mut cases, 10 (67%) showed RUNX1 mutations at MDS or AML stage. Multivariate analysis revealed that del(5q) and RUNX1 mutations were independent negative prognostic factors for overall survival, while blast count >5% was not. In conclusion, SF3B1mut MDS has a favorable prognosis independent of blast count if karyotype and RUNX1 mutations are considered.
© 2022. The Author(s).
Conflict of interest statement
CH, WK, and TH declare part ownership of Munich Leukemia Laboratory (MLL). SH, HS, MM, GH, and CB are employed by the MLL.
Figures

References
-
- Hasserjan RP, Orazi A, Brunning RD, Le Beau MM, Porwit A, Baumann I, et al. Myelodysplastic syndromes. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Myelodysplastic syndromes. 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2017:97–120.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

