Efficient and safe single-cell cloning of human pluripotent stem cells using the CEPT cocktail
- PMID: 36261632
- PMCID: PMC11009857
- DOI: 10.1038/s41596-022-00753-z
Efficient and safe single-cell cloning of human pluripotent stem cells using the CEPT cocktail
Abstract
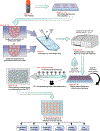
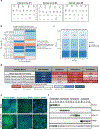
Human pluripotent stem cells (hPSCs) are inherently sensitive cells. Single-cell dissociation and the establishment of clonal cell lines have been long-standing challenges. This inefficiency of cell cloning represents a major obstacle for the standardization and streamlining of gene editing in induced pluripotent stem cells for basic and translational research. Here we describe a chemically defined protocol for robust single-cell cloning using microfluidics-based cell sorting in combination with the CEPT small-molecule cocktail. This advanced strategy promotes the viability and cell fitness of self-renewing stem cells. The use of low-pressure microfluidic cell dispensing ensures gentle and rapid dispensing of single cells into 96- and 384-well plates, while the fast-acting CEPT cocktail minimizes cellular stress and maintains cell structure and function immediately after cell dissociation. The protocol also facilitates clone picking and produces genetically stable clonal cell lines from hPSCs in a safe and cost-efficient fashion. Depending on the proliferation rate of the clone derived from a single cell, this protocol can be completed in 7-14 d and requires experience with aseptic cell culture techniques. Altogether, the relative ease, scalability and robustness of this workflow should boost gene editing in hPSCs and leverage a wide range of applications, including cell line development (e.g., reporter and isogenic cell lines), disease modeling and applications in regenerative medicine.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Ethics declarations
Competing interests
I.S., Y.C., and A.S. are co-inventors on a U.S. Department of Health and Human Services patent application covering the CEPT cocktail and its use.
Figures




References
-
- Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). - PubMed
-
- Takahashi K et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131, 861–872 (2007). - PubMed
-
- Evans MJ & Kaufman MH Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). - PubMed
Key references using this protocol:
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

