Biological roles of adenine methylation in RNA
- PMID: 36261710
- PMCID: PMC9974562
- DOI: 10.1038/s41576-022-00534-0
Biological roles of adenine methylation in RNA
Abstract
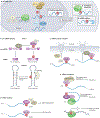
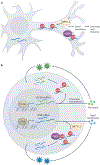
N6-Methyladenosine (m6A) is one of the most abundant modifications of the epitranscriptome and is found in cellular RNAs across all kingdoms of life. Advances in detection and mapping methods have improved our understanding of the effects of m6A on mRNA fate and ribosomal RNA function, and have uncovered novel functional roles in virtually every species of RNA. In this Review, we explore the latest studies revealing roles for m6A-modified RNAs in chromatin architecture, transcriptional regulation and genome stability. We also summarize m6A functions in biological processes such as stem-cell renewal and differentiation, brain function, immunity and cancer progression.
© 2022. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures



References
-
- Wei CM, Gershowitz A & Moss B Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386 (1975). - PubMed
-
- Sommer S, Lavi U & Darnell JE Jr. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol 124, 487–499 (1978). - PubMed
-
- Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). - PubMed
-
Together with Meyer et al. (2012), this paper reports the mapping of m6A in the human and mouse transcriptome.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

