Genome Evolution of a Symbiont Population for Pathogen Defense in Honeybees
- PMID: 36263788
- PMCID: PMC9648514
- DOI: 10.1093/gbe/evac153
Genome Evolution of a Symbiont Population for Pathogen Defense in Honeybees
Abstract
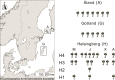
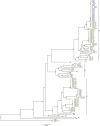
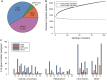
The honeybee gut microbiome is thought to be important for bee health, but the role of the individual members is poorly understood. Here, we present closed genomes and associated mobilomes of 102 Apilactobacillus kunkeei isolates obtained from the honey crop (foregut) of honeybees sampled from beehives in Helsingborg in the south of Sweden and from the islands Gotland and Åland in the Baltic Sea. Each beehive contained a unique composition of isolates and repeated sampling of similar isolates from two beehives in Helsingborg suggests that the bacterial community is stably maintained across bee generations during the summer months. The sampled bacterial population contained an open pan-genome structure with a high genomic density of transposons. A subset of strains affiliated with phylogroup A inhibited growth of the bee pathogen Melissococcus plutonius, all of which contained a 19.5 kb plasmid for the synthesis of the antimicrobial compound kunkecin A, while a subset of phylogroups B and C strains contained a 32.9 kb plasmid for the synthesis of a putative polyketide antibiotic. This study suggests that the mobile gene pool of A. kunkeei plays a key role in pathogen defense in honeybees, providing new insights into the evolutionary dynamics of defensive symbiont populations.
Keywords: Apilactobacillus kunkeei; defensive symbionts; evolution; mobile elements; plasmids; transposons.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures





References
-
- Anderson KE, Ricigliano VA. 2017. Honey bee gut dysbiosis: a novel context of disease ecology. Curr Opin Insect Sci. 22:125–132. - PubMed
-
- Arevalo P, VanInsberghe D, Elsherbini J, Gore J, Poltz MF. 2019. A reverse ecology approach based on biological definition of microbial populations. Cell 178:820–834. - PubMed
-
- Arredondo D, et al. . 2018. Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Benef Microbes. 9:279–290. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

