Specialized metabolism by trichome-enriched Rubisco and fatty acid synthase components
- PMID: 36264116
- PMCID: PMC9922422
- DOI: 10.1093/plphys/kiac487
Specialized metabolism by trichome-enriched Rubisco and fatty acid synthase components
Abstract
Acylsugars, specialized metabolites with defense activities, are secreted by trichomes of many solanaceous plants. Several acylsugar metabolic genes (AMGs) remain unknown. We previously reported multiple candidate AMGs. Here, using multiple approaches, we characterized additional AMGs. First, we identified differentially expressed genes between high- and low-acylsugar-producing F2 plants derived from a cross between cultivated tomato (Solanum lycopersicum) and a wild relative (Solanum pennellii), which produce acylsugars that are ∼1% and ∼20% of leaf dry weight, respectively. Expression levels of many known and candidate AMGs positively correlated with acylsugar amounts in F2 individuals. Next, we identified lycopersicum-pennellii putative orthologs with higher nonsynonymous to synonymous substitutions. These analyses identified four candidate genes, three of which showed enriched expression in stem trichomes compared to underlying tissues (shaved stems). Virus-induced gene silencing confirmed two candidates, Sopen05g009610 [beta-ketoacyl-(acyl-carrier-protein) reductase; fatty acid synthase component] and Sopen07g006810 (Rubisco small subunit), as AMGs. Phylogenetic analysis indicated that Sopen05g009610 is distinct from specialized metabolic cytosolic reductases but closely related to two capsaicinoid biosynthetic reductases, suggesting evolutionary relationship between acylsugar and capsaicinoid biosynthesis. Analysis of publicly available datasets revealed enriched expression of Sopen05g009610 orthologs in trichomes of several acylsugar-producing species. Similarly, orthologs of Sopen07g006810 were identified as solanaceous trichome-enriched members, which form a phylogenetic clade distinct from those of mesophyll-expressed "regular" Rubisco small subunits. Furthermore, δ13C analyses indicated recycling of metabolic CO2 into acylsugars by Sopen07g006810 and showed how trichomes support high levels of specialized metabolite production. These findings have implications for genetic manipulation of trichome-specialized metabolism in solanaceous crops.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures
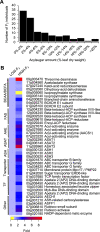



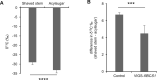
References
-
- Alba JM, Montserrat M, Fernandez-Munoz R (2009) Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Exp Appl Acarol 47: 35–47 - PubMed
-
- Asai T, Hara N, Fujimoto Y (2010) Fatty acid derivatives and dammarane triterpenes from the glandular trichome exudates of Ibicella lutea and Proboscidea louisiana. Phytochemistry 71: 877–894 - PubMed
-
- Asai T, Nakamura Y, Hirayama Y, Ohyama K, Fujimoto Y (2012) Cyclic glycolipids from glandular trichome exudates of Cerastium glomeratum. Phytochemistry 82: 149–157 - PubMed
-
- Balcke GU, Bennewitz S, Bergau N, Athmer B, Henning A, Majovsky P, Jiménez-Gómez JM, Hoehenwarter W, Tissier A (2017) Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 29: 960–983 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

