Profiling of cell-free DNA methylation and histone signatures in pediatric NAFLD: A pilot study
- PMID: 36264206
- PMCID: PMC9701487
- DOI: 10.1002/hep4.2082
Profiling of cell-free DNA methylation and histone signatures in pediatric NAFLD: A pilot study
Abstract
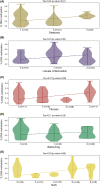
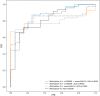
Nonalcoholic fatty liver disease (NAFLD) has become the most common chronic liver disease in children and adolescents, increasing the risk of its progression toward nonalcoholic steatohepatitis (NASH), cirrhosis, and cancer. There is an urgent need for noninvasive early diagnostic and prognostic tools such as epigenetic marks (epimarks), which would replace liver biopsy in the future. We used plasma samples from 67 children with biopsy-proven NAFLD, and as controls we used samples from 20 children negative for steatosis by ultrasound. All patients were genotyped for patatin-like phospholipase domain containing 3 (PNPLA3), transmembrane 6 superfamily member 2 (TM6SF2), membrane bound O-acyltransferase domain containing 7 (MBOAT7), and klotho-β (KLB) gene variants, and data on anthropometric and biochemical parameters were collected. Furthermore, plasma cell-free DNA (cfDNA) methylation was quantified using a commercially available kit, and ImageStream(X) was used for the detection of free circulating histone complexes and variants. We found a significant enrichment of the levels of histone macroH2A1.2 in the plasma of children with NAFLD compared to controls, and a strong correlation between cfDNA methylation levels and NASH. Receiver operating characteristic curve analysis demonstrated that combination of cfDNA methylation, PNPLA3 rs738409 variant, coupled with either high-density lipoprotein cholesterol or alanine aminotransferase levels can strongly predict the progression of pediatric NAFLD to NASH with area under the curve >0.87. Conclusion: Our pilot study combined epimarks and genetic and metabolic markers for a robust risk assessment of NAFLD development and progression in children, offering a promising noninvasive tool for the consistent diagnosis and prognosis of pediatric NAFLD. Further studies are necessary to identify their pathogenic origin and function.
© 2022 The Authors. Hepatology Communications published by Wiley Periodicals LLC on behalf of American Association for the Study of Liver Diseases.
Conflict of interest statement
Nothing to report.
Figures



Similar articles
-
The Membrane-bound O-Acyltransferase7 rs641738 Variant in Pediatric Nonalcoholic Fatty Liver Disease.J Pediatr Gastroenterol Nutr. 2018 Jul;67(1):69-74. doi: 10.1097/MPG.0000000000001979. J Pediatr Gastroenterol Nutr. 2018. PMID: 29601441
-
Validation of PNPLA3 polymorphisms as risk factor for NAFLD and liver fibrosis in an admixed population.Ann Hepatol. 2019 May-Jun;18(3):466-471. doi: 10.1016/j.aohep.2018.10.004. Epub 2019 Apr 18. Ann Hepatol. 2019. PMID: 31054980
-
Effect of PNPLA3 polymorphism on diagnostic performance of various noninvasive markers for diagnosing and staging nonalcoholic fatty liver disease.J Gastroenterol Hepatol. 2020 Jun;35(6):1057-1064. doi: 10.1111/jgh.14894. Epub 2019 Dec 9. J Gastroenterol Hepatol. 2020. PMID: 31677195
-
Diagnosis of non-alcoholic fatty liver disease (NAFLD).Diabetologia. 2016 Jun;59(6):1104-11. doi: 10.1007/s00125-016-3944-1. Epub 2016 Apr 18. Diabetologia. 2016. PMID: 27091184 Review.
-
The genetic backgrounds in nonalcoholic fatty liver disease.Clin J Gastroenterol. 2018 Apr;11(2):97-102. doi: 10.1007/s12328-018-0841-9. Epub 2018 Feb 28. Clin J Gastroenterol. 2018. PMID: 29492830 Review.
Cited by
-
Role of genetic variants and DNA methylation of lipid metabolism-related genes in metabolic dysfunction-associated steatotic liver disease.Front Physiol. 2025 Mar 17;16:1562848. doi: 10.3389/fphys.2025.1562848. eCollection 2025. Front Physiol. 2025. PMID: 40166716 Free PMC article. Review.
-
Mitochondrial GpC and CpG DNA Hypermethylation Cause Metabolic Stress-Induced Mitophagy and Cholestophagy.Int J Mol Sci. 2023 Nov 16;24(22):16412. doi: 10.3390/ijms242216412. Int J Mol Sci. 2023. PMID: 38003603 Free PMC article.
-
Extracellular Histones Profiles of Pediatric H3K27-Altered Diffuse Midline Glioma.Mol Diagn Ther. 2025 Jan;29(1):129-141. doi: 10.1007/s40291-024-00754-6. Epub 2024 Nov 8. Mol Diagn Ther. 2025. PMID: 39514166
-
Loss of PPARα function promotes epigenetic dysregulation of lipid homeostasis driving ferroptosis and pyroptosis lipotoxicity in metabolic dysfunction associated Steatotic liver disease (MASLD).Front Mol Med. 2024 Jan 8;3:1283170. doi: 10.3389/fmmed.2023.1283170. eCollection 2023. Front Mol Med. 2024. PMID: 39086681 Free PMC article.
-
Circulating Cell-Free DNA in Metabolic Diseases.J Endocr Soc. 2025 Jan 15;9(2):bvaf006. doi: 10.1210/jendso/bvaf006. eCollection 2025 Jan 6. J Endocr Soc. 2025. PMID: 39850787 Free PMC article. Review.
References
-
- Nobili V, Alisi A, Valenti L, Miele L, Feldstein AE, Alkhouri N. NAFLD in children: new genes, new diagnostic modalities and new drugs. Nat Rev Gastroenterol Hepatol. 2019;16:517–30. - PubMed
-
- Lazarus JV, Mark HE, Anstee QM, Arab JP, Batterham RL, Castera L, et al. Advancing the global public health agenda for NAFLD: a consensus statement. Nat Rev Gastroenterol Hepatol. 2022;19:60–78. - PubMed
-
- Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero‐Gomez M, et al. A new definition for metabolic dysfunction‐associated fatty liver disease: an international expert consensus statement. J Hepatol. 2020;73:202–9. - PubMed
-
- Eslam M, Alkhouri N, Vajro P, Baumann U, Weiss R, Socha P, et al. Defining paediatric metabolic (dysfunction)‐associated fatty liver disease: an international expert consensus statement. Lancet Gastroenterol Hepatol. 2021;6:864–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

