Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis
- PMID: 36265491
- PMCID: PMC9742301
- DOI: 10.1016/j.cmet.2022.09.026
Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis
Abstract
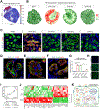
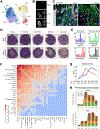
The underlying cellular events driving kidney fibrogenesis and metabolic dysfunction are incompletely understood. Here, we employed single-cell combinatorial indexing RNA sequencing to analyze 24 mouse kidneys from two fibrosis models. We profiled 309,666 cells in one experiment, representing 50 cell types/states encompassing epithelial, endothelial, immune, and stromal populations. Single-cell analysis identified diverse injury states of the proximal tubule, including two distinct early-phase populations with dysregulated lipid and amino acid metabolism, respectively. Lipid metabolism was defective in the chronic phase but was transiently activated in the very early stages of ischemia-induced injury, where we discovered increased lipid deposition and increased fatty acid β-oxidation. Perilipin 2 was identified as a surface marker of intracellular lipid droplets, and its knockdown in vitro disrupted cell energy state maintenance during lipid accumulation. Surveying epithelial cells across nephron segments identified shared and unique injury responses. Stromal cells exhibited high heterogeneity and contributed to fibrogenesis by epithelial-stromal crosstalk.
Keywords: PLIN2; acute kidney injury; cell differentiation; chronic kidney disease; fibrosis; kidney; lipid droplet; lipid metabolism; single-cell combinatorial indexing; tissue regeneration.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.D.H. is a consultant for Janssen Research & Development, LLC, Pfizer, and Chinook Therapeutics and holds equity in Chinook Therapeutics and grant funding from Chinook Therapeutics, Janssen Research & Development, LLC, and Pfizer; all interests are unrelated to the current work.
Figures





Comment in
-
Epithelial injury responses in kidney fibrosis.Nat Rev Nephrol. 2023 Jan;19(1):3. doi: 10.1038/s41581-022-00657-x. Nat Rev Nephrol. 2023. PMID: 36446960 No abstract available.
-
Stray tubules: the varied and dynamic fate of tubular cells.Kidney Int. 2023 Jul;104(1):19-21. doi: 10.1016/j.kint.2023.02.005. Epub 2023 Feb 18. Kidney Int. 2023. PMID: 36804410 No abstract available.
References
-
- Abe J. ichi, Ko KA, Kotla S, Wang Y, Paez-Mayorga J, Shin IJ, Imanishi M, Vu HT, Tao Y, Leiva-Juarez MM, Thomas TN, Medina JL, Won JH, Fujii Y, Giancursio CJ, McBeath E, Shin JH, Guzman L, Abe RJ, Taunton J, Mochizuki N, Faubion W, Cooke JP, Fujiwara K, Evans SE, Le NT, 2019. MAGI1 as a link between endothelial activation and ER stress drives atherosclerosis. JCI insight 4. 10.1172/JCI.INSIGHT.125570 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

