p53 mutation in normal esophagus promotes multiple stages of carcinogenesis but is constrained by clonal competition
- PMID: 36266286
- PMCID: PMC9584949
- DOI: 10.1038/s41467-022-33945-y
p53 mutation in normal esophagus promotes multiple stages of carcinogenesis but is constrained by clonal competition
Abstract
Aging normal human oesophagus accumulates TP53 mutant clones. These are the origin of most oesophageal squamous carcinomas, in which biallelic TP53 disruption is almost universal. However, how p53 mutant clones expand and contribute to cancer development is unclear. Here we show that inducing the p53R245W mutant in single oesophageal progenitor cells in transgenic mice confers a proliferative advantage and clonal expansion but does not disrupt normal epithelial structure. Loss of the remaining p53 allele in mutant cells results in genomically unstable p53R245W/null epithelium with giant polyaneuploid cells and copy number altered clones. In carcinogenesis, p53 mutation does not initiate tumour formation, but tumours developing from areas with p53 mutation and LOH are larger and show extensive chromosomal instability compared to lesions arising in wild type epithelium. We conclude that p53 has distinct functions at different stages of carcinogenesis and that LOH within p53 mutant clones in normal epithelium is a critical step in malignant transformation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

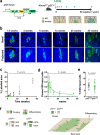

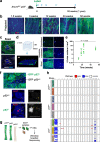
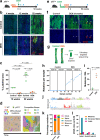


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

