Rapid antibiotic susceptibility testing and species identification for mixed samples
- PMID: 36266330
- PMCID: PMC9584937
- DOI: 10.1038/s41467-022-33659-1
Rapid antibiotic susceptibility testing and species identification for mixed samples
Abstract
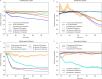
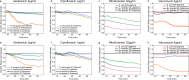
Antimicrobial resistance is an increasing problem on a global scale. Rapid antibiotic susceptibility testing (AST) is urgently needed in the clinic to enable personalized prescriptions in high-resistance environments and to limit the use of broad-spectrum drugs. Current rapid phenotypic AST methods do not include species identification (ID), leaving time-consuming plating or culturing as the only available option when ID is needed to make the sensitivity call. Here we describe a method to perform phenotypic AST at the single-cell level in a microfluidic chip that allows subsequent genotyping by in situ FISH. By stratifying the phenotypic AST response on the species of individual cells, it is possible to determine the susceptibility profile for each species in a mixed sample in 2 h. In this proof-of-principle study, we demonstrate the operation with four antibiotics and mixed samples with combinations of seven species.
© 2022. The Author(s).
Conflict of interest statement
J.E. has patented the method (US10,041,104) and founded the company Astrego Diagnostics. All other authors declare no competing interests.
Figures



References
-
- O’neill. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Review on antimicrobial resistance. https://amr-review.org/ (2014).
-
- O’Neil, J. Tackling Drug-resistant Infections Globally: Final Report and Recommendations. https://books.google.com/books/about/Tackling_Drug_resistant_Infections_... (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

