A fluorescent multi-domain protein reveals the unfolding mechanism of Hsp70
- PMID: 36266349
- PMCID: PMC9889267
- DOI: 10.1038/s41589-022-01162-9
A fluorescent multi-domain protein reveals the unfolding mechanism of Hsp70
Erratum in
-
Author Correction: A fluorescent multi-domain protein reveals the unfolding mechanism of Hsp70.Nat Chem Biol. 2023 Apr;19(4):529. doi: 10.1038/s41589-023-01296-4. Nat Chem Biol. 2023. PMID: 36849817 Free PMC article. No abstract available.
Abstract
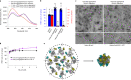
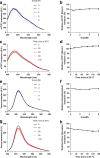
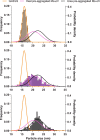
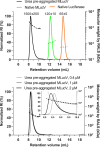
Detailed understanding of the mechanism by which Hsp70 chaperones protect cells against protein aggregation is hampered by the lack of a comprehensive characterization of the aggregates, which are typically heterogeneous. Here we designed a reporter chaperone substrate, MLucV, composed of a stress-labile luciferase flanked by stress-resistant fluorescent domains, which upon denaturation formed a discrete population of small aggregates. Combining Förster resonance energy transfer and enzymatic activity measurements provided unprecedented details on the aggregated, unfolded, Hsp70-bound and native MLucV conformations. The Hsp70 mechanism first involved ATP-fueled disaggregation and unfolding of the stable pre-aggregated substrate, which stretched MLucV beyond simply unfolded conformations, followed by native refolding. The ATP-fueled unfolding and refolding action of Hsp70 on MLucV aggregates could accumulate native MLucV species under elevated denaturing temperatures highly adverse to the native state. These results unambiguously exclude binding and preventing of aggregation from the non-equilibrium mechanism by which Hsp70 converts stable aggregates into metastable native proteins.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

