Nitric oxide-driven modifications of lipoic arm inhibit α-ketoacid dehydrogenases
- PMID: 36266351
- PMCID: PMC9974485
- DOI: 10.1038/s41589-022-01153-w
Nitric oxide-driven modifications of lipoic arm inhibit α-ketoacid dehydrogenases
Abstract
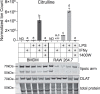
Pyruvate dehydrogenase complex (PDHC) and oxoglutarate dehydrogenase complex (OGDC), which belong to the mitochondrial α-ketoacid dehydrogenase family, play crucial roles in cellular metabolism. These multi-subunit enzyme complexes use lipoic arms covalently attached to their E2 subunits to transfer an acyl group to coenzyme A (CoA). Here, we report a novel mechanism capable of substantially inhibiting PDHC and OGDC: reactive nitrogen species (RNS) can covalently modify the thiols on their lipoic arms, generating a series of adducts that block catalytic activity. S-Nitroso-CoA, a product between RNS and the E2 subunit's natural substrate, CoA, can efficiently deliver these modifications onto the lipoic arm. We found RNS-mediated inhibition of PDHC and OGDC occurs during classical macrophage activation, driving significant rewiring of cellular metabolism over time. This work provides a new mechanistic link between RNS and mitochondrial metabolism with potential relevance for numerous physiological and pathological conditions in which RNS accumulate.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





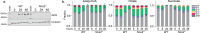

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

