Modeling gene × environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression
- PMID: 36266471
- PMCID: PMC9630117
- DOI: 10.1038/s41593-022-01161-y
Modeling gene × environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression
Erratum in
-
Author Correction: Modeling gene × environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression.Nat Neurosci. 2025 Jan;28(1):214. doi: 10.1038/s41593-024-01854-6. Nat Neurosci. 2025. PMID: 39633181 Free PMC article. No abstract available.
Abstract
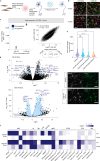
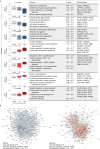
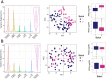
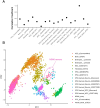
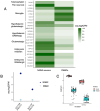
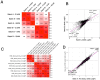
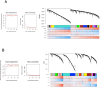
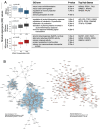
Post-traumatic stress disorder (PTSD) can develop following severe trauma, but the extent to which genetic and environmental risk factors contribute to individual clinical outcomes is unknown. Here, we compared transcriptional responses to hydrocortisone exposure in human induced pluripotent stem cell (hiPSC)-derived glutamatergic neurons and peripheral blood mononuclear cells (PBMCs) from combat veterans with PTSD (n = 19 hiPSC and n = 20 PBMC donors) and controls (n = 20 hiPSC and n = 20 PBMC donors). In neurons only, we observed diagnosis-specific glucocorticoid-induced changes in gene expression corresponding with PTSD-specific transcriptomic patterns found in human postmortem brains. We observed glucocorticoid hypersensitivity in PTSD neurons, and identified genes that contribute to this PTSD-dependent glucocorticoid response. We find evidence of a coregulated network of transcription factors that mediates glucocorticoid hyper-responsivity in PTSD. These findings suggest that induced neurons represent a platform for examining the molecular mechanisms underlying PTSD, identifying biomarkers of stress response, and conducting drug screening to identify new therapeutics.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
Modeling PTSD neuronal stress responses in a dish.Nat Neurosci. 2022 Nov;25(11):1402-1404. doi: 10.1038/s41593-022-01172-9. Nat Neurosci. 2022. PMID: 36266472 Free PMC article.
References
-
- Yehuda, R. et al. Post-traumatic stress disorder. Nat. Rev. Dis. Primers1, 15057 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01ES033630/U.S. Department of Health & Human Services | NIH | National Institute of Environmental Health Sciences (NIEHS)
- R01 MH118278/MH/NIMH NIH HHS/United States
- R01MH124839/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 MH124839/MH/NIMH NIH HHS/United States
- R01MH118278/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

