Expression of divergent methyl/alkyl coenzyme M reductases from uncultured archaea
- PMID: 36266535
- PMCID: PMC9584954
- DOI: 10.1038/s42003-022-04057-6
Expression of divergent methyl/alkyl coenzyme M reductases from uncultured archaea
Abstract
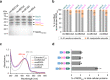
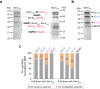
Methanogens and anaerobic methane-oxidizing archaea (ANME) are important players in the global carbon cycle. Methyl-coenzyme M reductase (MCR) is a key enzyme in methane metabolism, catalyzing the last step in methanogenesis and the first step in anaerobic methane oxidation. Divergent mcr and mcr-like genes have recently been identified in uncultured archaeal lineages. However, the assembly and biochemistry of MCRs from uncultured archaea remain largely unknown. Here we present an approach to study MCRs from uncultured archaea by heterologous expression in a methanogen, Methanococcus maripaludis. Promoter, operon structure, and temperature were important determinants for MCR production. Both recombinant methanococcal and ANME-2 MCR assembled with the host MCR forming hybrid complexes, whereas tested ANME-1 MCR and ethyl-coenzyme M reductase only formed homogenous complexes. Together with structural modeling, this suggests that ANME-2 and methanogen MCRs are structurally similar and their reaction directions are likely regulated by thermodynamics rather than intrinsic structural differences.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

