Identification of environmental factors that promote intestinal inflammation
- PMID: 36266581
- PMCID: PMC9898826
- DOI: 10.1038/s41586-022-05308-6
Identification of environmental factors that promote intestinal inflammation
Abstract
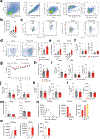
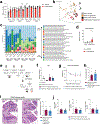
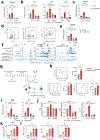
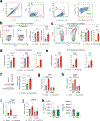
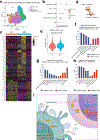
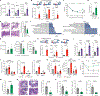
Genome-wide association studies have identified risk loci linked to inflammatory bowel disease (IBD)1-a complex chronic inflammatory disorder of the gastrointestinal tract. The increasing prevalence of IBD in industrialized countries and the augmented disease risk observed in migrants who move into areas of higher disease prevalence suggest that environmental factors are also important determinants of IBD susceptibility and severity2. However, the identification of environmental factors relevant to IBD and the mechanisms by which they influence disease has been hampered by the lack of platforms for their systematic investigation. Here we describe an integrated systems approach, combining publicly available databases, zebrafish chemical screens, machine learning and mouse preclinical models to identify environmental factors that control intestinal inflammation. This approach established that the herbicide propyzamide increases inflammation in the small and large intestine. Moreover, we show that an AHR-NF-κB-C/EBPβ signalling axis operates in T cells and dendritic cells to promote intestinal inflammation, and is targeted by propyzamide. In conclusion, we developed a pipeline for the identification of environmental factors and mechanisms of pathogenesis in IBD and, potentially, other inflammatory diseases.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures








Comment in
-
Zebrafishing for toxins.Nat Rev Immunol. 2022 Dec;22(12):718. doi: 10.1038/s41577-022-00801-5. Nat Rev Immunol. 2022. PMID: 36357705 No abstract available.
References
-
- Kamm MA Rapid changes in epidemiology of inflammatory bowel disease. Lancet 390, 2741–2742 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH130458/MH/NIMH NIH HHS/United States
- R01 ES032323/ES/NIEHS NIH HHS/United States
- R01 ES025530/ES/NIEHS NIH HHS/United States
- K99 NS114111/NS/NINDS NIH HHS/United States
- R01 AI126880/AI/NIAID NIH HHS/United States
- T32 GM007356/GM/NIGMS NIH HHS/United States
- T32 DK007356/DK/NIDDK NIH HHS/United States
- K12 CA090354/CA/NCI NIH HHS/United States
- R56 AI093903/AI/NIAID NIH HHS/United States
- R21 NS087867/NS/NINDS NIH HHS/United States
- R01 AI093903/AI/NIAID NIH HHS/United States
- R00 NS114111/NS/NINDS NIH HHS/United States
- F32 NS101790/NS/NINDS NIH HHS/United States
- T32 CA207021/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

