Comparative genomic and transcriptomic analyses of trans-kingdom pathogen Fusarium solani species complex reveal degrees of compartmentalization
- PMID: 36266645
- PMCID: PMC9583462
- DOI: 10.1186/s12915-022-01436-7
Comparative genomic and transcriptomic analyses of trans-kingdom pathogen Fusarium solani species complex reveal degrees of compartmentalization
Abstract
Background: The Fusarium solani species complex (FSSC) comprises fungal pathogens responsible for mortality in a diverse range of animals and plants, but their genome diversity and transcriptome responses in animal pathogenicity remain to be elucidated. We sequenced, assembled and annotated six chromosome-level FSSC clade 3 genomes of aquatic animal and plant host origins. We established a pathosystem and investigated the expression data of F. falciforme and F. keratoplasticum in Chinese softshell turtle (Pelodiscus sinensis) host.
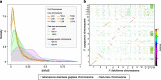
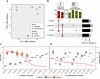
Results: Comparative analyses between the FSSC genomes revealed a spectrum of conservation patterns in chromosomes categorised into three compartments: core, fast-core (FC), and lineage-specific (LS). LS chromosomes contribute to variations in genomes size, with up to 42.2% of variations between F. vanettenii strains. Each chromosome compartment varied in structural architectures, with FC and LS chromosomes contain higher proportions of repetitive elements with genes enriched in functions related to pathogenicity and niche expansion. We identified differences in both selection in the coding sequences and DNA methylation levels between genome features and chromosome compartments which suggest a multi-speed evolution that can be traced back to the last common ancestor of Fusarium. We further demonstrated that F. falciforme and F. keratoplasticum are opportunistic pathogens by inoculating P. sinensis eggs and identified differentially expressed genes also associated with plant pathogenicity. These included the most upregulated genes encoding the CFEM (Common in Fungal Extracellular Membrane) domain.
Conclusions: The high-quality genome assemblies provided new insights into the evolution of FSSC chromosomes, which also serve as a resource for studies of fungal genome evolution and pathogenesis. This study also establishes an animal model for fungal pathogens of trans-kingdom hosts.
Keywords: Animal pathogenicity; Chromosome evolution; Fusarium solani species complex; Genome compartments; Opportunistic pathogen; Turtle.
© 2022. The Author(s).
Conflict of interest statement
Not applicable.
Figures





References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

